For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTKSGETTRKVLLEVRDAVVHYGRIRALHGVSLVVHEGELVTLLGSNGAGKTTMMRAISGLRPLTSGSVW FDGQDISRMKAHRRVTEGLIQAPEGRGVFPGMTVVENLEMGCYGRKFPSRQEHAERLDWVFETFPRLAER RSQVGGTLSGGEQQMLAIGRALMARPKILLLDEPSMGLAPMVISQIFRIIADINSQGTTVLLVEQNAQQA LSRSDRAYILETGNITRTGVARELLQDDSIRAAYLGVA
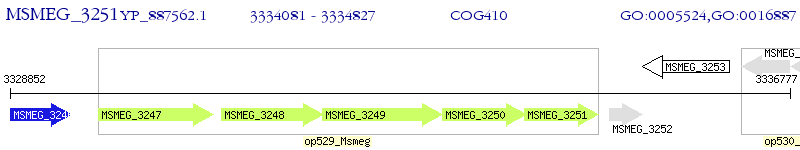
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3251 | - | - | 100% (248) | branched-chain amino acid ABC transporter ATP-binding protein |
| M. smegmatis MC2 155 | MSMEG_1217 | - | 1e-43 | 37.78% (225) | ABC-type transport system ATP-binding protein I |
| M. smegmatis MC2 155 | MSMEG_2978 | - | 2e-41 | 42.06% (233) | ABC transporter ATP-binding protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2419c | cysA1 | 2e-23 | 32.87% (216) | sulfate-transport ATP-binding protein ABC transporter CysA1 |
| M. gilvum PYR-GCK | Mflv_3570 | - | 1e-119 | 86.29% (248) | ABC transporter related |
| M. tuberculosis H37Rv | Rv2397c | cysA1 | 2e-23 | 32.87% (216) | sulfate-transport ATP-binding protein ABC transporter CysA1 |
| M. leprae Br4923 | MLBr_00590 | - | 7e-23 | 29.82% (228) | putative ABC transporter ATP-binding protein |
| M. abscessus ATCC 19977 | MAB_2622c | - | 3e-99 | 74.68% (237) | branched chain amino acid ABC transporter |
| M. marinum M | MMAR_3715 | cysA1 | 1e-24 | 32.76% (232) | sulfate-transport ATP-binding protein ABC transporter CysA1 |
| M. avium 104 | MAV_1785 | - | 2e-24 | 32.34% (235) | sulfate/thiosulfate import ATP-binding protein CysA |
| M. thermoresistible (build 8) | TH_3858 | - | 1e-117 | 88.24% (238) | PUTATIVE ABC transporter related |
| M. ulcerans Agy99 | MUL_3658 | cysA1 | 1e-24 | 32.76% (232) | sulfate-transport ATP-binding protein ABC transporter CysA1 |
| M. vanbaalenii PYR-1 | Mvan_2845 | - | 1e-119 | 88.33% (240) | ABC transporter-related protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2419c|M.bovis_AF2122/97 ------MT--------YAIVVADATKRYGDFVALDHVDFVVPTGSLTALL
Rv2397c|M.tuberculosis_H37Rv ------MT--------YAIVVADATKRYGDFVALDHVDFVVPTGSLTALL
MMAR_3715|M.marinum_M MKKETSLTGPGT--GGHAIVVRDAFKQYGDFVALDHVDFVVPAGSLTALL
MUL_3658|M.ulcerans_Agy99 ------MTGPGT--GGHAIVVRDAFKQYGDFVALDHVDFVVPAGSLTALL
MAV_1785|M.avium_104 ------MTDAGTDRGDIAITVRDAYKRYGDFVALDHVDFVVPTGSLTALL
MSMEG_3251|M.smegmatis_MC2_155 -MTKS-----GETTRKVLLEVRDAVVHYGRIRALHGVSLVVHEGELVTLL
TH_3858|M.thermoresistible__bu ----------------VMLEVRDVEVHYGRIRALHGVSLRVHEGELVTLL
Mflv_3570|M.gilvum_PYR-GCK -MTNS------PDPRPVLLEVRDAVVHYGRIKALHSVSLTVHEGELVTLL
Mvan_2845|M.vanbaalenii_PYR-1 -MSSPETVAGQPDPRPVLLEVRDVVVHYGRIKALHSVSLTVHEGELVTLL
MAB_2622c|M.abscessus_ATCC_199 -------------MPPVQLKLNNVGVHYGPIQAVNTLSFDVREGELVTLL
MLBr_00590|M.leprae_Br4923 ----MSSACTVPETPEVVIRLRGVCKHYGSILAVSNLDLDIHSAEVLALL
: : .. :** : *: :.: : ..: :**
Mb2419c|M.bovis_AF2122/97 GPSGSGKSTLLRTIAGLDQPDTGTITINGRDVTRVPPQRR---GIGFVFQ
Rv2397c|M.tuberculosis_H37Rv GPSGSGKSTLLRTIAGLDQPDTGTITINGRDVTRVPPQRR---GIGFVFQ
MMAR_3715|M.marinum_M GPSGSGKSTLLRTIAGLDQPDSGNITINGRDVTRVPPQRR---GIGFVFQ
MUL_3658|M.ulcerans_Agy99 GPSGSGKSTLLRTIAGLDQPDSGNITINGRDVTRVPPQRR---GIGFVFQ
MAV_1785|M.avium_104 GPSGSGKSTLLRTIAGLDQPDTGTVTIYGRDVTRVPPQRR---GIGFVFQ
MSMEG_3251|M.smegmatis_MC2_155 GSNGAGKTTMMRAISGLRPLTSGSVWFDGQDISRMKAHRRVTEGLIQAPE
TH_3858|M.thermoresistible__bu GSNGAGKTTMMRAISGLRPLTRGSVWFDGRDITRVSAHRRVADGLIQVPE
Mflv_3570|M.gilvum_PYR-GCK GSNGAGKTTMMRAISGLLPLTSGSVWFEGKDISRVKAHKRVTDGLIQAPE
Mvan_2845|M.vanbaalenii_PYR-1 GSNGAGKTTMMRAISGLLPLSSGSVWFDGKDISRVKAHKRVADGLIQAPE
MAB_2622c|M.abscessus_ATCC_199 GSNGAGKSTAMRAISGLVPLSSGTIFFEGQDISKMKAHRRVRAGIVQAPE
MLBr_00590|M.leprae_Br4923 GPNGAGKTTTVEMCEGFVRPDAGTIEVLG--LDPITDNARLRARIGVMLQ
*..*:**:* :. *: *.: . * : : : * : :
Mb2419c|M.bovis_AF2122/97 HYAAFKHLTVRDNVAFGLKIRKRP-KAEIKAKVDNLLQVVG-LSGFQSRY
Rv2397c|M.tuberculosis_H37Rv HYAAFKHLTVRDNVAFGLKIRKRP-KAEIKAKVDNLLQVVG-LSGFQSRY
MMAR_3715|M.marinum_M HYAAFKHLTVRDNVAYGLKIRKRP-KAEIRDKVDNLLEVVG-LSGFHGRY
MUL_3658|M.ulcerans_Agy99 HYAAFKHLTVRDNVAYGLKIRKRP-KAEIRDKVDNLLEVVG-LSGFHGRY
MAV_1785|M.avium_104 HYAAFKHLTVRDNVAYGLKVRKRP-KAEIKAKVDNLLEVVG-LSGFQGRY
MSMEG_3251|M.smegmatis_MC2_155 GRGVFPGMTVVENLEMGCYGRKFPSRQEHAERLDWVFETFPRLAERRSQV
TH_3858|M.thermoresistible__bu GRGVFPGMTVLENLDMGCYGRRFASRAEHRERLDWVLQTFPRLAERRDQV
Mflv_3570|M.gilvum_PYR-GCK GRGVFPGMTIIDNLEMGCYGRKFATNAEHDEKLDWVLETFPRLAERRSQV
Mvan_2845|M.vanbaalenii_PYR-1 GRGVFPGMTIVENLEMGCYGRKFPSKAEHDERLDWVLETFPRLAERRNQV
MAB_2622c|M.abscessus_ATCC_199 GRGIFPGMTVAENLEMGCYGRKFKSRAERRERLDWINALFPRLAERRHQP
MLBr_00590|M.leprae_Br4923 GGGGYPAARAGEMLNLVASYAADP------LDPEWLLETLG-LTDVARTT
. : : : : : . *:
Mb2419c|M.bovis_AF2122/97 PNQLSGGQRQRMALARALAVDPEVLLLDEPFGALDAKVREELRAWLRRLH
Rv2397c|M.tuberculosis_H37Rv PNQLSGGQRQRMALARALAVDPEVLLLDEPFGALDAKVREELRAWLRRLH
MMAR_3715|M.marinum_M PNQLSGGQRQRMALARALAVDPEVLLLDEPFGALDAKVREDLRAWLRRLH
MUL_3658|M.ulcerans_Agy99 PDQLSGGQRQRMALARALAVDPEVLLLDEPFGALDAKVREDLRAWLRRLH
MAV_1785|M.avium_104 PNQLSGGQRQRMALARALAVDPQVLLLDEPFGALDAKVREDLRAWLRRLH
MSMEG_3251|M.smegmatis_MC2_155 GGTLSGGEQQMLAIGRALMARPKILLLDEPSMGLAPMVISQIFRIIADIN
TH_3858|M.thermoresistible__bu GGTLSGGEQQMLAIGRALMARPKVLLLDEPSMGLAPMVISQIFGIIADIN
Mflv_3570|M.gilvum_PYR-GCK GGTLSGGEQQMLAIGRALMARPKVLLLDEPSMGLAPMVISQIFKIIAEIN
Mvan_2845|M.vanbaalenii_PYR-1 GGTLSGGEQQMLAIGRALMARPKVLLLDEPSMGLAPMVISQIFRIIAEIN
MAB_2622c|M.abscessus_ATCC_199 GGTLSGGEQQMLSIGRALMARPKILLLDEPSMGLAPLLITKIFNIIEEIN
MLBr_00590|M.leprae_Br4923 YRRLSGGQQQRLALACTLVGRPELVFLDEPTAGMDTHARLLVWELIDTLR
****::* :::. :* *::::**** .: . : : :.
Mb2419c|M.bovis_AF2122/97 DEVHVTTVLVTHDQAEALDVADRIAVLHKGRIEQVGSPTDVYDAPANAFV
Rv2397c|M.tuberculosis_H37Rv DEVHVTTVLVTHDQAEALDVADRIAVLHKGRIEQVGSPTDVYDAPANAFV
MMAR_3715|M.marinum_M DEVHVTTVLVTHDQAEALDVADRIAVLNSGRIEQVGSPTEVYDAPANAFV
MUL_3658|M.ulcerans_Agy99 DEVHVTTVLVTHDQAEALDVADRIAVLNSGRIEQVGSPTEVYDAPTNAFV
MAV_1785|M.avium_104 DEVHVTTVLVTHDQAEALDVADRIAVLNQGRIEQIGSPTEVYDAPTNAFV
MSMEG_3251|M.smegmatis_MC2_155 -SQGTTVLLVEQNAQQALSRSDRAYILETGNITRTGVARELLQD--DSIR
TH_3858|M.thermoresistible__bu -AQGTTVLLVEQNAQQALSRSDRAYILETGEITRSGPARELLAD--DSIR
Mflv_3570|M.gilvum_PYR-GCK -ATGTTVLLVEQNAQQALSRSDRAYILETGEVTRTGNARELLAD--DSIR
Mvan_2845|M.vanbaalenii_PYR-1 -AAGTTVLLVEQNAQQALTRSDRAYILETGEVTRTGNARELLSD--KSIR
MAB_2622c|M.abscessus_ATCC_199 -AAGTTVVLVEQNAQQALSRSDHAYILETGSVVRSGAGRELLQD--DSIR
MLBr_00590|M.leprae_Br4923 -RDGVTVVLTTHQLKEAEELADRLVIIDHGRMIAAGTPTELMHTGAKDQL
.*.:*. :: :* :*: ::. * : * :: .
Mb2419c|M.bovis_AF2122/97 MSFLGAVSTLNGSLVRPHDIRVGRTPNMAVAAADGTAGSTGVLRAVVDRV
Rv2397c|M.tuberculosis_H37Rv MSFLGAVSTLNGSLVRPHDIRVGRTPNMAVAAADGTAGSTGVLRAVVDRV
MMAR_3715|M.marinum_M MSFLGAVSALNGALVRPHDIRVGRTPEMAVAAADGTGESTGVTRAVVDRI
MUL_3658|M.ulcerans_Agy99 MSFLGAVSALNGALVRPHDIRVGRTPEMAVAAADGTGESTGVTRAVVDRI
MAV_1785|M.avium_104 MSFLGAVSTLNGTLVRPHDIRVGRTPEMAVAAEDGTAESTGVARAIVDRV
MSMEG_3251|M.smegmatis_MC2_155 AAYLGVA-------------------------------------------
TH_3858|M.thermoresistible__bu AAYLGVA-------------------------------------------
Mflv_3570|M.gilvum_PYR-GCK AAYLGVA-------------------------------------------
Mvan_2845|M.vanbaalenii_PYR-1 AAYLGVA-------------------------------------------
MAB_2622c|M.abscessus_ATCC_199 SAYLGVV-------------------------------------------
MLBr_00590|M.leprae_Br4923 RFSAPPRLDLSLLAFALPEDYKTTELTPGEYLVAGPIDPQVLATVTAWCA
Mb2419c|M.bovis_AF2122/97 VVLGFEVRVELTSAATGGAFTAQITRGDAEALALREGDTVYVRATRVPPI
Rv2397c|M.tuberculosis_H37Rv VVLGFEVRVELTSAATGGAFTAQITRGDAEALALREGDTVYVRATRVPPI
MMAR_3715|M.marinum_M VVLGFEVRVELTSAATGGAFTAQITRGDAEALALQEGDTVYVRATRVPPI
MUL_3658|M.ulcerans_Agy99 VVLGFEVRVELTSAATGGAFTAQITRGDAEALALQEGDTVYVRATRVPPI
MAV_1785|M.avium_104 VKLGFEVRVELTSAATGGPFTAQITRGDAEALALREGDTVYVRATRVPPI
MSMEG_3251|M.smegmatis_MC2_155 --------------------------------------------------
TH_3858|M.thermoresistible__bu --------------------------------------------------
Mflv_3570|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2845|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_2622c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_00590|M.leprae_Br4923 QINVLTTNMRVEQHSLEDVFLDLTGRKLR---------------------
Mb2419c|M.bovis_AF2122/97 AG---GVSGVDDAGVERVKVTST------
Rv2397c|M.tuberculosis_H37Rv AG---GVSGVDDAGVERVKVTST------
MMAR_3715|M.marinum_M AG---AAPAVPDDGAGSTPSAAQIGVGGQ
MUL_3658|M.ulcerans_Agy99 AG---AAPAVPDDGAGSTPRAAQIGVGGQ
MAV_1785|M.avium_104 TAGATTVPAVSRDGADEATLTSA------
MSMEG_3251|M.smegmatis_MC2_155 -----------------------------
TH_3858|M.thermoresistible__bu -----------------------------
Mflv_3570|M.gilvum_PYR-GCK -----------------------------
Mvan_2845|M.vanbaalenii_PYR-1 -----------------------------
MAB_2622c|M.abscessus_ATCC_199 -----------------------------
MLBr_00590|M.leprae_Br4923 -----------------------------