For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSDQHTQQNPGENTSSLVRTLLAPGDIIREQWSTLPRVAKWAVGVIGFGLLALLPLFTPGFLNTPGISFG GTMAQFAMVAIIAIGLNVVVGQAGLLDLGYVGFYAVGAYTVALLTSPSSPWNIVGVDKFLSSDWAWLACV PLAMAITAIAGLILGTPTLRLRGDYLAIVTLGFGEIIRLMADNLSDITNGARGLNQVAYPRLGESEKLPN GVFSSGNSTGVANYGTWWFWLGLALIVGILLLVGNLERSRVGRAWIAIREDEDAAEVMGVNAFKFKLWAF TIGAAIGGLSGALYAGQVQYVAPQTFNIINSMLFLCAVVLGGQGNKLGVIFGAFVIVYLPNRLLGVEFLG INLGDLKYLFFGLALVVLMIFRPQGLFPARQHLLTYGKAARDLLRATPVKEHTS
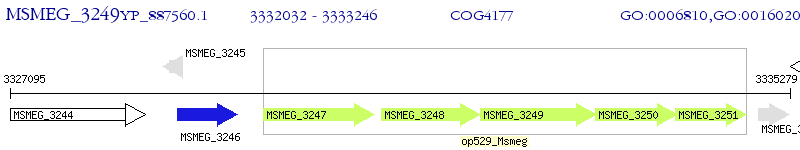
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3249 | - | - | 100% (404) | branched-chain amino acid ABC transporter, permease protein |
| M. smegmatis MC2 155 | MSMEG_6878 | - | 1e-30 | 28.87% (336) | inner-membrane translocator |
| M. smegmatis MC2 155 | MSMEG_1219 | - | 7e-25 | 26.57% (335) | ABC-type transport system permease protein II |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3572 | - | 1e-178 | 82.75% (371) | inner-membrane translocator |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2624c | - | 1e-163 | 74.93% (375) | high-affinity branched-chain amino acid ABC transporter (LivM) |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_3840 | - | 0.0 | 83.73% (381) | PUTATIVE branched-chain amino acid ABC transporter, permease |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_2843 | - | 1e-180 | 83.56% (371) | inner-membrane translocator |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3572|M.gilvum_PYR-GCK ----------------MTR--------VMEWWDGLTRAQKWVFGIVLFAG
Mvan_2843|M.vanbaalenii_PYR-1 ---MSTVEKAG---AVWAR--------VMDWWDGLTRAQKWVFGIILFAG
MSMEG_3249|M.smegmatis_MC2_155 MSDQHTQQNPGENTSSLVRTLLAPGDIIREQWSTLPRVAKWAVGVIGFGL
TH_3840|M.thermoresistible__bu ---VNRLLAPG----DVVR----------RWWDSRTRGHKWAVGLLGFGV
MAB_2624c|M.abscessus_ATCC_199 -MSAARALAPG----DSIR----------RWWSELARPVQWVYGVVGFAL
* *. .* :*. *:: *.
Mflv_3572|M.gilvum_PYR-GCK IALSPLFKPGFIDTPGISFGGTMAQFAMVAIIAIGLNVVVGQAGLLDLGY
Mvan_2843|M.vanbaalenii_PYR-1 IALSPLFTPGFIDTPGISFGGTMAQFAMVAIIAIGLNVVVGQAGLLDLGY
MSMEG_3249|M.smegmatis_MC2_155 LALLPLFTPGFLNTPGISFGGTMAQFAMVAIIAIGLNVVVGQAGLLDLGY
TH_3840|M.thermoresistible__bu LALLPLYTPPFLDTPGISFGGTMAQFAMFALIAIGLNVVVGQAGLLDLGY
MAB_2624c|M.abscessus_ATCC_199 LALLPVWKIPYLDTPNTSFGGVMAQFAMVALIAIGLNVVVGQAGLLDLGY
:** *::. :::**. ****.******.*:*******************
Mflv_3572|M.gilvum_PYR-GCK VGFYAVGAYTVALLTSPESPWNQMSPKGILTTPWAWLACVPLAMAVTALS
Mvan_2843|M.vanbaalenii_PYR-1 VGFYAVGAYTVALLTSPESPWNKLGPTGFFSTPWAWLSCVPLAMAVTALS
MSMEG_3249|M.smegmatis_MC2_155 VGFYAVGAYTVALLTSPSSPWNIVGVDKFLSSDWAWLACVPLAMAITAIA
TH_3840|M.thermoresistible__bu VGFYAVGAYTVALLTSPESPWNRMGWTVFFSSDWAWLSCVPLAMAVTAFS
MAB_2624c|M.abscessus_ATCC_199 VGFYAIGAYTVALLTSPDSPWNRMGTSAFFSSDWAWLSCLPIAVAITACF
*****:***********.**** :. :::: ****:*:*:*:*:**
Mflv_3572|M.gilvum_PYR-GCK GLILGTPTLRLRGDYLAIVTLGFGEIIRLLADNLADVTNGPRGLNEVAFP
Mvan_2843|M.vanbaalenii_PYR-1 GLILGTPTLRLRGDYLAIVTLGFGEIIRLLADNLADITNGPRGLNEVAFP
MSMEG_3249|M.smegmatis_MC2_155 GLILGTPTLRLRGDYLAIVTLGFGEIIRLMADNLSDITNGARGLNQVAYP
TH_3840|M.thermoresistible__bu GLIIGTPTLRLRGDYLAIVTLGFGEIIRLVADNLSGVTNGARGLHQVAYP
MAB_2624c|M.abscessus_ATCC_199 GLILGTPTLRLRGDYLAIVTLGFGEIIRLLADNL-DVTNGPRGLHKVAYP
***:*************************:**** .:***.***::**:*
Mflv_3572|M.gilvum_PYR-GCK HFLESEQHPEGVFSVSNSGGDANYGAWWFWLGLILIVGILLLVGNLERSR
Mvan_2843|M.vanbaalenii_PYR-1 HFLESDQHPEGVFSVSNSGGDANYGTWWFWLGLILIVGILLLVGNLERSR
MSMEG_3249|M.smegmatis_MC2_155 RLGESEKLPNGVFSSGNSTGVANYGTWWFWLGLALIVGILLLVGNLERSR
TH_3840|M.thermoresistible__bu RVGESERLPDGVFSSGNSAGAANYGTWWFWLGLVLIAVILVLVGNLERSR
MAB_2624c|M.abscessus_ATCC_199 HVGEQHVHGGGVFSSANSGGHLNYGVWWYWLGLCLIAAVLVLMGNLERSR
:. *.. **** .** * ***.**:**** **. :*:*:*******
Mflv_3572|M.gilvum_PYR-GCK VGRAWIAVREDEDAAEVMGVNAFKFKLWAFTIGAAIGGLSGALYAGQVQY
Mvan_2843|M.vanbaalenii_PYR-1 VGRAWIAVREDEDAAEVMGVNAFKFKLWAFTIGAAIGGLSGALYAGQVQY
MSMEG_3249|M.smegmatis_MC2_155 VGRAWIAIREDEDAAEVMGVNAFKFKLWAFTIGAAIGGLSGALYAGQVQY
TH_3840|M.thermoresistible__bu VGRAWIAIREDEDAAEVMGVNTFKFKVWAFVIGAAIGGLSGALYAGQVQY
MAB_2624c|M.abscessus_ATCC_199 VGRSWVAIREDEDAAEMMGVPTFRFKLWAFVIGASIGGLSGALYAGQVQY
***:*:*:********:*** :*:**:***.***:***************
Mflv_3572|M.gilvum_PYR-GCK VAPPTFNIINSMLFLCAVVLGGQGNKLGVILGAFIIVYLPNRLLGVHFLG
Mvan_2843|M.vanbaalenii_PYR-1 VAPPTFNIINSMLFLCAVVLGGQGNKLGVILGAFIIVYLPNRLLGVHFLG
MSMEG_3249|M.smegmatis_MC2_155 VAPQTFNIINSMLFLCAVVLGGQGNKLGVIFGAFVIVYLPNRLLGVEFLG
TH_3840|M.thermoresistible__bu VAPPTFNIINSMLFLCAVVLGGQGNKLGVVLGAFVIVYLPNRLLGVELLG
MAB_2624c|M.abscessus_ATCC_199 VASPTFNIINSMLFLCAVVLGGQGNKLGVVVGAFIIVYLPNRLLGVQVQG
**. *************************:.***:***********.. *
Mflv_3572|M.gilvum_PYR-GCK IDMGNLKYLFFGLALVVLMIFRPQGLFPARQHLLTYARAARKLLRAKPTD
Mvan_2843|M.vanbaalenii_PYR-1 IDMGNLKYLFFGLALVVLMIFRPQGLFPARQHLLTYARAARKLLRAQPTD
MSMEG_3249|M.smegmatis_MC2_155 INLGDLKYLFFGLALVVLMIFRPQGLFPARQHLLTYGKAARDLLRATPVK
TH_3840|M.thermoresistible__bu VNLGDLKYLFFGLALVVMMIFRPAGLFPARQQLLAYGRSAREMLRARPDD
MAB_2624c|M.abscessus_ATCC_199 IDLGNLKYLFFGLALVVLMLFRPQGLFPAHQKLMAYWRKTSERLSAQKPD
:::*:************:*:*** *****:*:*::* : : . * * .
Mflv_3572|M.gilvum_PYR-GCK TEPAK-
Mvan_2843|M.vanbaalenii_PYR-1 TEPAK-
MSMEG_3249|M.smegmatis_MC2_155 EHTS--
TH_3840|M.thermoresistible__bu REPVSS
MAB_2624c|M.abscessus_ATCC_199 EVEAGA