For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TKHQRLTSAVAASIVATAGVAVSQAPAPASTGTPTAHQTPTSTSTPRWLTVTSTPEGAPATPNALYGAIL PSTSPGSVTSTGSGIGHREPGISGSPAFTAVRAVRPGSSTARLVFAPKTSTSGTALTVTPIAVGMVGNAK VTVAVDSSGRPMIGPGGWLIGNGIEPGQNGGLLIGNGADGGPGQKGGNGGFLLGNGGRGGDGLAGVNGGR AGDGGNAGLLGNGGDGGNSVGGVEGANGGNAGLIGNGGKGGDGSPNVISNGAHIANAGRGGNGGNGGLLV GSGGDGGRGGDGDGEGPETAGSHGADGGNGGAGGVFGNGGRGGDGGDANNGTVGTRNGNAAAGKGGNGGS TGIGTGGAGGNGGKATGTNAAYGGNGGNGGSGLAGGGNGGDGGDAQSSSNRARPGQGGQAGRGGFGRDGK TGNPGNATDRSATRENVSNRVTGRSTPTRSADKAGSRTSSSSESNSESNSESSSKTSSKAGGTKSSDTKS GGGAAGTGSDSKDSE*
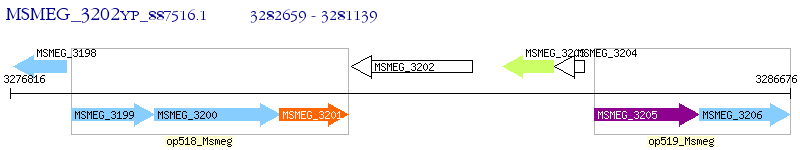
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3202 | - | - | 100% (506) | hypothetical protein MSMEG_3202 |
| M. smegmatis MC2 155 | MSMEG_0868 | - | 5e-52 | 39.19% (370) | hypothetical protein MSMEG_0868 |
| M. smegmatis MC2 155 | MSMEG_0076 | - | 3e-10 | 29.27% (205) | antigen MTB48 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2648c | PE_PGRS45 | 3e-62 | 48.88% (313) | PE-PGRS family protein |
| M. gilvum PYR-GCK | Mflv_5091 | - | 1e-58 | 34.50% (516) | triple helix repeat-containing collagen |
| M. tuberculosis H37Rv | Rv2615c | PE_PGRS45 | 5e-63 | 48.59% (319) | PE-PGRS family protein |
| M. leprae Br4923 | MLBr_01556 | infB | 7e-10 | 26.46% (257) | translation initiation factor IF-2 |
| M. abscessus ATCC 19977 | MAB_4284c | - | 2e-31 | 30.23% (440) | hypothetical protein MAB_4284c |
| M. marinum M | MMAR_0806 | - | 3e-61 | 40.15% (391) | PE-PGRS family protein |
| M. avium 104 | MAV_4827 | - | 1e-19 | 41.88% (160) | hypothetical protein MAV_4827 |
| M. thermoresistible (build 8) | TH_3810 | - | 2e-38 | 36.89% (347) | PUTATIVE - |
| M. ulcerans Agy99 | MUL_3318 | - | 5e-64 | 40.60% (399) | PE-PGRS family protein |
| M. vanbaalenii PYR-1 | Mvan_3867 | - | 6e-74 | 37.78% (532) | PE-PGRS family protein |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_3202|M.smegmatis_MC2_155 -MTKHQRLTSAVAASIVATAGVAVSQAPAPASTGTPTAHQTPTSTSTPRW
Mvan_3867|M.vanbaalenii_PYR-1 --MPHPILATAMAVGIAATAGAGALGDAPPTVN--VTANKHVSATRLTAV
Mb2648c|M.bovis_AF2122/97 -------MSF---VNVAPQLVSTAAADAARIGSAINTANTAAA-ATTQVL
Rv2615c|M.tuberculosis_H37Rv -------MSF---VNVAPQLVSTAAADAARIGSAINTANTAAA-ATTQVL
MMAR_0806|M.marinum_M -------MSF---VSVVPEWVAAAATDVAGIGSVVGAANAAAAGATTSVT
MUL_3318|M.ulcerans_Agy99 -------MSY---LAIAPEIVAAAAADMDGIGSPITAANAAAAIPTSAIA
Mflv_5091|M.gilvum_PYR-GCK ------MVTFGGGATPTPTSLGAAAVDLSGTSCGLICNGQDGTGQNGGIL
TH_3810|M.thermoresistible__bu ------------------------------------VDGDGGSGGRTGLF
MAB_4284c|M.abscessus_ATCC_199 MSIIDFILNLFRDPVEAASYVADPEAALANAGLSAVSAAQVGAVMPVVAE
MAV_4827|M.avium_104 -------MNIVLGVSMAPSSIQMVVLEGEYADGATVEEDAIDVTGADDAA
MLBr_01556|M.leprae_Br4923 --MAGKARVHELAKELGVTSKEVLARLNEQGEFVKSASSTVEAPVARRLR
MSMEG_3202|M.smegmatis_MC2_155 LTVT--------STPEGAPATPNALYGAI----LPSTSPGSVTSTG--SG
Mvan_3867|M.vanbaalenii_PYR-1 VTPN--------ALYSAAPASDVVVLGAHG--SLAVGEPRGVPQAGALAG
Mb2648c|M.bovis_AF2122/97 VAAQ-----------DEVSTAIAALFGSHG----QHYQAISAQVAAYQER
Rv2615c|M.tuberculosis_H37Rv AAAQ-----------DEVSTAIAALFGSHG----QHYQAISAQVAAYQQR
MMAR_0806|M.marinum_M AAAG-----------DEVSVAIAALFGGFG----REYQAVCGQWAEFEQR
MUL_3318|M.ulcerans_Agy99 AAAA-----------DEISAAIAELFGNHA----QQYQALSAQMARFHDQ
Mflv_5091|M.gilvum_PYR-GCK FGNGGSGWNSDVVGMDGGNGGNGGLFGGNGGNGGNGLGGGNGGNGGNAGA
TH_3810|M.thermoresistible__bu SKLS-----------RGGDGGDGGLGGNGG----------HGGGTGIISL
MAB_4284c|M.abscessus_ATCC_199 SVASNYGYQSQWVNTGNPIHDLSGNLQNYYAAQPDGPSILSPSLLSPRDN
MAV_4827|M.avium_104 TASAPDRVISAILGTREGAADAGLQLSSIG----VTWTDQLEAAALRDAL
MLBr_01556|M.leprae_Br4923 ESFGGIKPAADKGAEQVATKAQAKRLGESLDQTLDRALDKAVAGNGATTA
MSMEG_3202|M.smegmatis_MC2_155 IGHREPG---ISGSPAFTAVRAVRPGSSTARLVFAPKTSTSGTALTVTPI
Mvan_3867|M.vanbaalenii_PYR-1 ASRAVPGGLELSTGLATSRVHPL-PERVNTDIEMVSAPTTSGPALRLGRA
Mb2648c|M.bovis_AF2122/97 FVLALSQAGSTYAVAEAASATPL-------QNVLDAINAP-----VQSLT
Rv2615c|M.tuberculosis_H37Rv FVLALSQAGSTYAVAEAASATPL-------QNVLDAINAP-----VQSLT
MMAR_0806|M.marinum_M FARALGAGAGAYAGAEAAAVGTLSPLGVIDQGVWGAVNGP-----AVALW
MUL_3318|M.ulcerans_Agy99 FVRSLSGAAQTYARAEATGAAPL-------QPVLDLINGP-----TQALL
Mflv_5091|M.gilvum_PYR-GCK FALFGNGGNGGNGGVGVVGATGAVG----APGVAGATGATGVTSGVVGGN
TH_3810|M.thermoresistible__bu LSLAGNGGNG----------------------------------------
MAB_4284c|M.abscessus_ATCC_199 DFLSHNDTDFASHNPIASGNQVMSPSGNVDSFNRGPLLDLSFGDLQFGNR
MAV_4827|M.avium_104 AARRIDNVMLVSAFLAAAALAQNVGGAMGYERTAVLLVEPDTATLAVVET
MLBr_01556|M.leprae_Br4923 APVQVDHSAAVVPIVAGEGPSTAHREELAPPAGQPSEQPGVPLPGQQGTP
MSMEG_3202|M.smegmatis_MC2_155 AVGMVGNAKVTVAVDSSGRPMIGPGGWLIGNGIEPGQNGGLLIG------
Mvan_3867|M.vanbaalenii_PYR-1 TTPSVG------STGPAGAAAPGLFSLLIGNGDEPGENGGWLIG------
Mb2648c|M.bovis_AF2122/97 GRPLIGDGANG---IDGTGQAGGNGGWLWGNGGNSGSGAPGQAG------
Rv2615c|M.tuberculosis_H37Rv GRPLIGDGANG---IDGTGQAGGNGGWLWGNGGNGGSGAPGQAG------
MMAR_0806|M.marinum_M GRPLIGDGADG---AAGTGQAGGAGGLLWGNGGNGGSGGVGQNG------
MUL_3318|M.ulcerans_Agy99 GRPLIGNGTSG---ALGTGQAGGDGGLLIGNG-TSGALGTGQAG------
Mflv_5091|M.gilvum_PYR-GCK GEDAVGYGATG---AAGAAGAAGAAGAAGAVGSAGAAGEAGETGTW----
TH_3810|M.thermoresistible__bu -----GNGYRT---TDNNALDGGHG----GNGGNVGFFSLFNLG------
MAB_4284c|M.abscessus_ATCC_199 DTTAIGDGAVAVGGSNSGPIASGQGSTAVNGDVRDSNIINADHG------
MAV_4827|M.avium_104 SDGSIPDVYKQQIRAESYGDAAGQLSAMVAGLEKLESLPGGLLLVG----
MLBr_01556|M.leprae_Br4923 AAPHPGHPGMPTGPHPGPAPKPGGRPPRVGNNPFSSAQSVARPIPRPPAP
*
MSMEG_3202|M.smegmatis_MC2_155 ---NGADGGPGQKGGNG---------------------------------
Mvan_3867|M.vanbaalenii_PYR-1 ---NGADGGPGQDGGRG---------------------------------
Mb2648c|M.bovis_AF2122/97 -GAGGAAGLIGN-GGAG---------------------------------
Rv2615c|M.tuberculosis_H37Rv -GAGGAAGLIGN-GGAG---------------------------------
MMAR_0806|M.marinum_M -GAGGSAGLIGR-GGAGGVGGLSGASGAGGAGGVGGNGGWLWGAGGDGGA
MUL_3318|M.ulcerans_Agy99 -GDGG--LLIGN-GGAG---------------------------------
Mflv_5091|M.gilvum_PYR-GCK -GNAGLPGKAGDAGGAGAVG------------------------------
TH_3810|M.thermoresistible__bu -GAGG-------WGGSG---------------------------------
MAB_4284c|M.abscessus_ATCC_199 -SNVGVDQSQTRVGGDYTNVSGHG--------------------------
MAV_4827|M.avium_104 -SGVDVAPLKQVLQTTTFLDVS----------------------------
MLBr_01556|M.leprae_Br4923 RPSASPSSMSPRPGGAVGGGGP----------------------------
.
MSMEG_3202|M.smegmatis_MC2_155 -------------------------------GFLLGNGGRGG--------
Mvan_3867|M.vanbaalenii_PYR-1 -------------------------------GLLFGNGGRGG--------
Mb2648c|M.bovis_AF2122/97 --------------------------GAGGQGLPFEAGANGG--------
Rv2615c|M.tuberculosis_H37Rv --------------------------GAGGQGLPFEAGANGG--------
MMAR_0806|M.marinum_M GGVGALSGGVGAAAGRAWLWGAGGVGGAGGQGMGLDGGAGGGGGQGGLVY
MUL_3318|M.ulcerans_Agy99 -----------------------------GSGAAGQAGGAGG--------
Mflv_5091|M.gilvum_PYR-GCK --------------------------AAGAAGTVGTAGATGGTGTG----
TH_3810|M.thermoresistible__bu -----------------------------VDGVDVVGGNPDG--------
MAB_4284c|M.abscessus_ATCC_199 -------------------------SADIDKSVTVVDDHSQRNTTN----
MAV_4827|M.avium_104 --------------------------VAEEPETALARGAALAAANAP---
MLBr_01556|M.leprae_Br4923 --------------------------RPPRTGVPRPGGGRPGAPVGG---
.
MSMEG_3202|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_3867|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb2648c|M.bovis_AF2122/97 -----------------------------AGGAG----------------
Rv2615c|M.tuberculosis_H37Rv -----------------------------AGGAG----------------
MMAR_0806|M.marinum_M GGGGDGGAGGVGALSGGVGGAAGRAWLWGAGGAGGAGGQGMGLDGGAGGG
MUL_3318|M.ulcerans_Agy99 -----------------------------AGGAAG---------------
Mflv_5091|M.gilvum_PYR-GCK ----------------------------ANGGAGG---------------
TH_3810|M.thermoresistible__bu --------------------------------------------------
MAB_4284c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_4827|M.avium_104 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 --------------------------------------------------
MSMEG_3202|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_3867|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb2648c|M.bovis_AF2122/97 --------------------------------------------------
Rv2615c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0806|M.marinum_M GGQGGLIYGGGGDGGAGGVGGEVGGGGAGGTGGAGGAAGLIGHGGAGGGG
MUL_3318|M.ulcerans_Agy99 --------------------------------------------------
Mflv_5091|M.gilvum_PYR-GCK --------------------------------------------------
TH_3810|M.thermoresistible__bu --------------------------------------------------
MAB_4284c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_4827|M.avium_104 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 --------------------------------------------------
MSMEG_3202|M.smegmatis_MC2_155 ----------------------------------DGLAGVNGGRAGDGGN
Mvan_3867|M.vanbaalenii_PYR-1 ----------------------------------DALT--SGGPAGRGGD
Mb2648c|M.bovis_AF2122/97 ------------------------------GWLFGNGG---AGGNGGIGG
Rv2615c|M.tuberculosis_H37Rv ------------------------------GWLFGNGG---AGGNGGIGG
MMAR_0806|M.marinum_M GAGDGGSTGGAGQTGGDGGRAGDGGVGGRGGWLAGAGGDGGAGGAGGVGG
MUL_3318|M.ulcerans_Agy99 --------------------------------LIGSGG---AGGIGGTGA
Mflv_5091|M.gilvum_PYR-GCK --------------------------------VGGVGGTGAAGAAGGAGG
TH_3810|M.thermoresistible__bu ---------------------------------------------GTPGA
MAB_4284c|M.abscessus_ATCC_199 --------------------------------INDSFNTDRSTNIGSGNT
MAV_4827|M.avium_104 ------------------------------LFASSTAALAYAQDPGTGAV
MLBr_01556|M.leprae_Br4923 ---------------------------------RSDAGGGNYRGGGVGAL
*
MSMEG_3202|M.smegmatis_MC2_155 AG--------LLGNGGDGGN----SVGGVEGANGGN--------------
Mvan_3867|M.vanbaalenii_PYR-1 AG--------LFGNGGDGGTGFVSSAAGTNAASGGTG-------------
Mb2648c|M.bovis_AF2122/97 AG----TNLAIGGHGGNGGNAGLIGAGGTGGAGGTG--------------
Rv2615c|M.tuberculosis_H37Rv AG----TNLAIGGHGGNGGNAGLIGAGGTGGAGGTG--------------
MMAR_0806|M.marinum_M AGGGGADGLVLGSDGGDGGDGGTGGAGGTGGAGGAGGLISFFGGQGAGGA
MUL_3318|M.ulcerans_Agy99 AG-------------GKGGNAVLFGAGGNGGIGGTG--------------
Mflv_5091|M.gilvum_PYR-GCK LG-------ETGTTGGAGGVGGQGTAGGTGGVGGIG--------------
TH_3810|M.thermoresistible__bu HG-------------ESHTPGVYTDRQGEDGEDGTG--------------
MAB_4284c|M.abscessus_ATCC_199 TN-------TEIGSHNQANIGTSTSAGGSAAVGGNAG-------------
MAV_4827|M.avium_104 DQYSLPDYLYVPVDPEPGDDELAYSAVGEEDAESPTVVIEKLFLP-----
MLBr_01556|M.leprae_Br4923 PGGGSGGFRGRPGGGGHGGGGRPGQRGGAAGAFGRPG-------------
* .
MSMEG_3202|M.smegmatis_MC2_155 ---------------------------------------AG---------
Mvan_3867|M.vanbaalenii_PYR-1 --------------------------------------GAGGNGGL----
Mb2648c|M.bovis_AF2122/97 ----------------------------------GGEPSAGASG------
Rv2615c|M.tuberculosis_H37Rv ----------------------------------GGEPSAGASG------
MMAR_0806|M.marinum_M GGAGGAGGLAGDGGVGATGTFAGGGSGTGGAGGDGGTPGVGGAGGAGGAG
MUL_3318|M.ulcerans_Agy99 --------------------------------------AVGGTG------
Mflv_5091|M.gilvum_PYR-GCK -----------------------------------GAGSTGAVG------
TH_3810|M.thermoresistible__bu ------------------------------------------TG------
MAB_4284c|M.abscessus_ATCC_199 --------------------------------------LAGNAG------
MAV_4827|M.avium_104 -------------------------------------EDSEPRRRP----
MLBr_01556|M.leprae_Br4923 -----------------------------------GAPRRGRKSKR----
MSMEG_3202|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_3867|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb2648c|M.bovis_AF2122/97 --------------------------------------------------
Rv2615c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0806|M.marinum_M SIAGAHGSEGARPLSGNGGSGGRGADSTVPGGIAGDGGVGGNGGVFGNGG
MUL_3318|M.ulcerans_Agy99 --------------------------------------------------
Mflv_5091|M.gilvum_PYR-GCK --------------------------------------------------
TH_3810|M.thermoresistible__bu --------------------------------------------------
MAB_4284c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_4827|M.avium_104 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 --------------------------------------------------
MSMEG_3202|M.smegmatis_MC2_155 -------------------------------LIGNGGKGGDGSPNVISNG
Mvan_3867|M.vanbaalenii_PYR-1 -------------------------------LMGDGGKGGTGGRAISAEG
Mb2648c|M.bovis_AF2122/97 ---------------------------------GNGGNGGNGG--LLIGN
Rv2615c|M.tuberculosis_H37Rv ---------------------------------GNGGNGGNGG--LLIGN
MMAR_0806|M.marinum_M TGGAGGTGIAGQRGVSADTPGGSGTAGEDGGVGGNGGAGGLGG--ALAGH
MUL_3318|M.ulcerans_Agy99 ---------------------------------AVGGAGGNGG--FLFGH
Mflv_5091|M.gilvum_PYR-GCK ---------------------------------GVGGKGIQGGQGLQGGK
TH_3810|M.thermoresistible__bu ----------------------------------IGGRGGNGG-----SS
MAB_4284c|M.abscessus_ATCC_199 -------------------------------LAGNAGLAGNAGLAGNAGL
MAV_4827|M.avium_104 -----------------------------VLLVGSGLAVVGISAVLALEI
MLBr_01556|M.leprae_Br4923 ------------------QKRQEYDSMQAPVVGGVRLPHGNGETIRLARG
MSMEG_3202|M.smegmatis_MC2_155 -AHIANAGRGGN--------------------------------------
Mvan_3867|M.vanbaalenii_PYR-1 PANGGNGGRGGNAGV-----------------------------------
Mb2648c|M.bovis_AF2122/97 SGDGGAAGNG----------------------------------------
Rv2615c|M.tuberculosis_H37Rv SGDGGAAGNG----------------------------------------
MMAR_0806|M.marinum_M GGDGGAGGTGGDGGAGGSGAAGTAGDRGTIAHLDGGRGGDGGSGADGGAG
MUL_3318|M.ulcerans_Agy99 GGDGGTGGTG----------------------------------------
Mflv_5091|M.gilvum_PYR-GCK GGDGGAGGKGGD--------------------------------------
TH_3810|M.thermoresistible__bu SGTHPMAEDG----------------------------------------
MAB_4284c|M.abscessus_ATCC_199 AGNAGLAGNAG---------------------------------------
MAV_4827|M.avium_104 ALAIGIRTTG----------------------------------------
MLBr_01556|M.leprae_Br4923 ASLSDFAEKIDANPAALVQALFN---------------------------
MSMEG_3202|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_3867|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb2648c|M.bovis_AF2122/97 --------------------------------------------------
Rv2615c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0806|M.marinum_M GAGGNGGAGGQALAAGYTDGVRGAGGSGGAGGAGGLAGAGGDGGDAYTTG
MUL_3318|M.ulcerans_Agy99 --------------------------------------------------
Mflv_5091|M.gilvum_PYR-GCK --------------------------------------------------
TH_3810|M.thermoresistible__bu --------------------------------------------------
MAB_4284c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_4827|M.avium_104 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 --------------------------------------------------
MSMEG_3202|M.smegmatis_MC2_155 ------------------------------------GGNGG---------
Mvan_3867|M.vanbaalenii_PYR-1 -----------------------------LGGDAGGGGDGGRGYSE----
Mb2648c|M.bovis_AF2122/97 ------------------------------AGISQNGPASG---------
Rv2615c|M.tuberculosis_H37Rv ------------------------------AGISQNGPASG---------
MMAR_0806|M.marinum_M GGAGGDGGHGGDTGTGGAGGSGGAGSSNGLSGLSGHSPTSGGDGGAGGDG
MUL_3318|M.ulcerans_Agy99 -----------------------------------ASAAAG---------
Mflv_5091|M.gilvum_PYR-GCK -----------------------------ADSMGSAGATGG---------
TH_3810|M.thermoresistible__bu ----------------------------------EHVKALD---------
MAB_4284c|M.abscessus_ATCC_199 -----------------------------LAGNAGLAGNAG---------
MAV_4827|M.avium_104 ------------------------------TVALQPTPNQHLIVP-----
MLBr_01556|M.leprae_Br4923 -----------------------------LGEMVTATQSVGDETLELLG-
MSMEG_3202|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_3867|M.vanbaalenii_PYR-1 -------------------------------------KGPASGGAGGAGA
Mb2648c|M.bovis_AF2122/97 -------------------------------------FGGNGGHAGTTGL
Rv2615c|M.tuberculosis_H37Rv -------------------------------------FGGNGGHAGTTGL
MMAR_0806|M.marinum_M GDSPATGGRGGAGGNGGKYGNGGAGGAGGDGIARGDAAGGDGGNGGKGGF
MUL_3318|M.ulcerans_Agy99 ---------------------------------------ANGG-AGQTGL
Mflv_5091|M.gilvum_PYR-GCK -------------------------------------KGGNGGNGGRGGL
TH_3810|M.thermoresistible__bu ---------------------------------------------GRPGL
MAB_4284c|M.abscessus_ATCC_199 -------------------------------------LAGNAGLAGNAGL
MAV_4827|M.avium_104 ------------------------------------TEQAPAPPPVQASA
MLBr_01556|M.leprae_Br4923 ------------------------------SEMNYNVQVVSPEDEDRELL
MSMEG_3202|M.smegmatis_MC2_155 --LLVGSGGDGGRGG-----------------------------------
Mvan_3867|M.vanbaalenii_PYR-1 AASTVGNGGSGGKGG-----------------------------------
Mb2648c|M.bovis_AF2122/97 IG-NGGNGGAGGAGG-----------------------------------
Rv2615c|M.tuberculosis_H37Rv IG-NGGNGGAGGAGG-----------------------------------
MMAR_0806|M.marinum_M YG-DGGDGGAGGAGGSGTTGVGFEGGRGGAGGDGGAGGAYGDGGDGGAGG
MUL_3318|M.ulcerans_Agy99 AGTDGGTGGASGEGG-----------------------------------
Mflv_5091|M.gilvum_PYR-GCK FG-KGGNAGNGADGG-----------------------------------
TH_3810|M.thermoresistible__bu DG---GNGSHGGNGG-----------------------------------
MAB_4284c|M.abscessus_ATCC_199 AG-NAGLAGNAGLAG-----------------------------------
MAV_4827|M.avium_104 PQPKISVPEPVAAPKP----------------------------------
MLBr_01556|M.leprae_Br4923 EPFDLTYGEDQGDED-----------------------------------
.
MSMEG_3202|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_3867|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb2648c|M.bovis_AF2122/97 --------------------------------------------------
Rv2615c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0806|M.marinum_M AGGDGRNGVDATTAGASGTQGFAGGGGGAGGAGGAGGTQAGAGGAGGDGG
MUL_3318|M.ulcerans_Agy99 --------------------------------------------------
Mflv_5091|M.gilvum_PYR-GCK --------------------------------------------------
TH_3810|M.thermoresistible__bu --------------------------------------------------
MAB_4284c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_4827|M.avium_104 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 --------------------------------------------------
MSMEG_3202|M.smegmatis_MC2_155 --DGDGEGP-----------------------------------------
Mvan_3867|M.vanbaalenii_PYR-1 --SAQGAGS-----------------------------------------
Mb2648c|M.bovis_AF2122/97 -DVSADFGG-----------------------------------------
Rv2615c|M.tuberculosis_H37Rv -DVSADFGG-----------------------------------------
MMAR_0806|M.marinum_M AGGTGGFGGSGANGGAGDHGTAANPNGGRGGDGGSGAAGGAGGDGGNGGA
MUL_3318|M.ulcerans_Agy99 ---AGGKGG-----------------------------------------
Mflv_5091|M.gilvum_PYR-GCK ---TGGAGG-----------------------------------------
TH_3810|M.thermoresistible__bu ---HGGRGG-----------------------------------------
MAB_4284c|M.abscessus_ATCC_199 ---NAGLAG-----------------------------------------
MAV_4827|M.avium_104 --LSPPVAAP----------------------------------------
MLBr_01556|M.leprae_Br4923 ---ELQVRPP----------------------------------------
MSMEG_3202|M.smegmatis_MC2_155 ----------------------------------------ETAGSHG---
Mvan_3867|M.vanbaalenii_PYR-1 ----------------------------------------ANGGAGGSGG
Mb2648c|M.bovis_AF2122/97 ---------------------------------------VGFGGQGGNGG
Rv2615c|M.tuberculosis_H37Rv ---------------------------------------VGFGGQGGNGG
MMAR_0806|M.marinum_M GGQAQAAGYADGTRGAGGNGGVGGAGGLAGDGGRGGDGAVGFGGAGGDGG
MUL_3318|M.ulcerans_Agy99 ---------------------------------------LLFG-AGGEGG
Mflv_5091|M.gilvum_PYR-GCK -----------------------------------------AGGTGGTGG
TH_3810|M.thermoresistible__bu -----------------------------------------FLGIGGSGG
MAB_4284c|M.abscessus_ATCC_199 -----------------------------------------NAGLAGNAG
MAV_4827|M.avium_104 -----------------------------------------LPAAPPPAA
MLBr_01556|M.leprae_Br4923 -----------------------------------------VVTVMGHVD
MSMEG_3202|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_3867|M.vanbaalenii_PYR-1 AG------------------------------------------------
Mb2648c|M.bovis_AF2122/97 AG------------------------------------------------
Rv2615c|M.tuberculosis_H37Rv AG------------------------------------------------
MMAR_0806|M.marinum_M HGGNTGAGGAGGTGGVGSSTGLTGLGGHSPTSGGDGGNGGNGGRGAVDIA
MUL_3318|M.ulcerans_Agy99 AG------------------------------------------------
Mflv_5091|M.gilvum_PYR-GCK AG------------------------------------------------
TH_3810|M.thermoresistible__bu WG------------------------------------------------
MAB_4284c|M.abscessus_ATCC_199 LA------------------------------------------------
MAV_4827|M.avium_104 PP------------------------------------------------
MLBr_01556|M.leprae_Br4923 HG------------------------------------------------
MSMEG_3202|M.smegmatis_MC2_155 -------------ADGGNGGAGG-------------------------VF
Mvan_3867|M.vanbaalenii_PYR-1 ---------GFVSGDGGAGGGGGTA-------------------LSAAGT
Mb2648c|M.bovis_AF2122/97 ---------GLLYGNGGAGGNGG---------------------AAGSPG
Rv2615c|M.tuberculosis_H37Rv ---------GLLYGNGGAGGNGG---------------------AAGSPG
MMAR_0806|M.marinum_M GGAGGTGGNGGAYGNGGDGGDGGDGVNGTRGLTAITPGGSGTDGSAGSAG
MUL_3318|M.ulcerans_Agy99 -------------GTGGAGGTGG---------------------AGGDGL
Mflv_5091|M.gilvum_PYR-GCK -------------GTGGTGGTGG----------------------IGGAG
TH_3810|M.thermoresistible__bu ----------------GHGGNGG---------------------------
MAB_4284c|M.abscessus_ATCC_199 -------------GNAGLAGNAG------------------------LAG
MAV_4827|M.avium_104 -----------LPEAPPIPAAPPVVP------------------VPVVVP
MLBr_01556|M.leprae_Br4923 ---KTRLLDTIRKANVREAEAGG-----------------ITQHIGAYQV
MSMEG_3202|M.smegmatis_MC2_155 GNGGRGGDG-------------GDANNGTVGTRNGNAAAGK---------
Mvan_3867|M.vanbaalenii_PYR-1 GSGGSGGDG-------------GNARGAGGGGRGGVGALGQS--------
Mb2648c|M.bovis_AF2122/97 SVTAFGGNG-------------GSGGSGGNGGNALIGNAG----------
Rv2615c|M.tuberculosis_H37Rv SVTAFGGNG-------------GSGGSGGNGGNALIGNAG----------
MMAR_0806|M.marinum_M GAGGYGGNGGALAGDGGDGGDGGNGGSGGDGGSGANGGAGDNGTTTSPNG
MUL_3318|M.ulcerans_Agy99 AATTAGGTG-------------GNAGDGGEGGAG--GNAG----------
Mflv_5091|M.gilvum_PYR-GCK STGGTGGTG-------------GTGGTGGTGGTGGTGGTG----------
TH_3810|M.thermoresistible__bu DTDADGGNG----------------GDGGNGGSAGRGGTG----------
MAB_4284c|M.abscessus_ATCC_199 NAGLAGNAG-----------LAGNAGLAGNAAVAGNAAVAG---------
MAV_4827|M.avium_104 PVPVPVPVRIPVPDPVSPPQLLAPPRLSVPQPVQPPVRVPQ---------
MLBr_01556|M.leprae_Br4923 GVDLDGSER--------LITFIDTPGHEAFTAMRARGAKAT---------
MSMEG_3202|M.smegmatis_MC2_155 ------------------------------------------GGNGGSTG
Mvan_3867|M.vanbaalenii_PYR-1 ---------------------------------KDARGFGGVGGAGGRSG
Mb2648c|M.bovis_AF2122/97 --------------------------------------AGGSAGAGGNGA
Rv2615c|M.tuberculosis_H37Rv --------------------------------------AGGSAGAGGNGA
MMAR_0806|M.marinum_M GRGGDGGHGGHGGDGGAGGHGGVAGKAQAAGYTDGTHGAGGVGGTGGNGG
MUL_3318|M.ulcerans_Agy99 --------------------------------------AGGAGGQGGLFG
Mflv_5091|M.gilvum_PYR-GCK -------------------------------------GTGGTGGKGGTGG
TH_3810|M.thermoresistible__bu ----------------------------------------GLRGVFGING
MAB_4284c|M.abscessus_ATCC_199 --------------------------------------NAAVAGNAAVGG
MAV_4827|M.avium_104 --------------------------------------PPSPPQVGGTVP
MLBr_01556|M.leprae_Br4923 --------------------------------------DIAILVVAADDG
.
MSMEG_3202|M.smegmatis_MC2_155 IG------------TGGAGGNGGK--------------------------
Mvan_3867|M.vanbaalenii_PYR-1 VTG-----------TGGAGGAGGSG-------------------------
Mb2648c|M.bovis_AF2122/97 SAG----------TAGGSGGDGGKG-------------------------
Rv2615c|M.tuberculosis_H37Rv SAG----------TAGGSGGDGGKG-------------------------
MMAR_0806|M.marinum_M LAGDGGRGGNGAATIGGNGGDGGNGGYPGVGGAGGKGGIGETIGLSGLGG
MUL_3318|M.ulcerans_Agy99 NAG-----------TTGAGGDGGNG-------------------------
Mflv_5091|M.gilvum_PYR-GCK TGG-----------AGGTGGDGGTG-------------------------
TH_3810|M.thermoresistible__bu GNG-----------RAGLGGRGXXX-------------------------
MAB_4284c|M.abscessus_ATCC_199 VLN-----------TGVSGAIGGSS-------------------------
MAV_4827|M.avium_104 QSPPQHQTPPGEGTPPKPDGPGNYG-------------------------
MLBr_01556|M.leprae_Br4923 VMP---------QTVEAINHAQAADVP-----------------------
.
MSMEG_3202|M.smegmatis_MC2_155 ---------------------------------ATGTNAAYGGNGGNGGS
Mvan_3867|M.vanbaalenii_PYR-1 ---------------------------------VSDTLDAAGGSGGDGGD
Mb2648c|M.bovis_AF2122/97 ---------------------------------GNGGSVGLIGNGGNGGN
Rv2615c|M.tuberculosis_H37Rv ---------------------------------GNGGSVGLIGNGGNGGN
MMAR_0806|M.marinum_M YSPISGNGGDGGDGGSGSAGTTAHPAGFDGGAGGHGGDGGAFGDGGNGGD
MUL_3318|M.ulcerans_Agy99 ---------------------------------GNGGLAGNGGAGGNGDA
Mflv_5091|M.gilvum_PYR-GCK ---------------------------------GTGGTGGTGGTGGTGGT
TH_3810|M.thermoresistible__bu ----------------------------------XGGGVGAAVRSGNGVF
MAB_4284c|M.abscessus_ATCC_199 -------------------------------HVGGAINESLGGSGSVGGS
MAV_4827|M.avium_104 --------------------------------GGHVPTPGAGGSGGSGGG
MLBr_01556|M.leprae_Br4923 ------------------------------IVVAVNKIDKEGADPAKIRG
.
MSMEG_3202|M.smegmatis_MC2_155 GLAGGG----------------------NGGDGGDAQSSSNRARPGQGGQ
Mvan_3867|M.vanbaalenii_PYR-1 GSRGG-----------------------IGGTGGAAHSKSGNAQGGFAGD
Mb2648c|M.bovis_AF2122/97 GGNG-------------------------GAGSLFNGAPGFGGPGGSG--
Rv2615c|M.tuberculosis_H37Rv G----------------------------GAGSLFNGAPGFGGPGGSG--
MMAR_0806|M.marinum_M GGTGGTGTAATTAGSPGAVGGHGGAGGNGGDGGYLNGNAGNGGSGGAGGT
MUL_3318|M.ulcerans_Agy99 SNPN-------------------------GGTGGNAANPGAGGAGGAGG-
Mflv_5091|M.gilvum_PYR-GCK GGAG----------------------DGTGGTGGTGGTGGQGGQGGQGGT
TH_3810|M.thermoresistible__bu SGGG------------------------GTPSAGFGVPTTIQFPAGSCTS
MAB_4284c|M.abscessus_ATCC_199 TAVGGG--------------------AHTGVGGGVSTGVGVGSHVGGGIS
MAV_4827|M.avium_104 HVPTPG------------------AGGSGGSGGGHVPTPGTGGPGGTGGS
MLBr_01556|M.leprae_Br4923 QLTEYG--------------------LVAEDFGGDTMFIDISAKVGTNIE
. *
MSMEG_3202|M.smegmatis_MC2_155 AGRG--GFGRDGKTGNPGNATDRSATRENVSNRVTG-RSTPTRSADKAGS
Mvan_3867|M.vanbaalenii_PYR-1 GGTGSRGSGGDGGDGGIGTSTTGKGRGGNGGNGGGGIRAAPVGKAAKAR-
Mb2648c|M.bovis_AF2122/97 ------GASLLGPPGLAGTNGADG--------------------------
Rv2615c|M.tuberculosis_H37Rv ------GASLLGPPGLAGTNGADG--------------------------
MMAR_0806|M.marinum_M GGAGATGTSQAAATGLGGGRGGDGGAGGAGGDGGNGGKAHATGFHNGTGG
MUL_3318|M.ulcerans_Agy99 ------NGSRTGAPGTTGNTPTTAAG--HGGKGGDGFNPATSGQDGGSGG
Mflv_5091|M.gilvum_PYR-GCK G-----GTGGQGGLGGGGGTGGQGGTGGTGGQGGTGGTGGTGG-TGGTGG
TH_3810|M.thermoresistible__bu R-----AVSREGIMPSSRAASARAGAACASSTSRSRRSFCCSRA------
MAB_4284c|M.abscessus_ATCC_199 ESLGGSGSVGGSTAVGGGAHTGVGGGVSTGVGVGSHVGGGISESLGGSGS
MAV_4827|M.avium_104 GGGHVPTPGAGTPGGSGGGHLPFPGAGSTPGGSGGGHLPTPGAGTPGGGL
MLBr_01556|M.leprae_Br4923 ALLEAVLLTADAALDLRANSGMEAQGVAIEAHLDRGRGPVATVLVQRGTL
MSMEG_3202|M.smegmatis_MC2_155 RTSSSSESNSESNSESSSKTSSKAGGTKSSDTKSGGGAAGTGSDSKDSE-
Mvan_3867|M.vanbaalenii_PYR-1 RAAAVAEATAPAPDGVVSLGPAYAQSRSATNALCCNEFSCAVTNSSEW--
Mb2648c|M.bovis_AF2122/97 --------------------------------------------------
Rv2615c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0806|M.marinum_M QGGDGGQGGKAGNGGNGADGQAAAIGRPRVSPGDGGDGGNGGSGGTGGTG
MUL_3318|M.ulcerans_Agy99 KGGDAGQFGNGGNGGNGAAGDAAG---------SGHTGGNGGAGGGGGNA
Mflv_5091|M.gilvum_PYR-GCK AGGDGGLGGLGGFGGSGGTGGQGGLGGP-----GGSTGGAGGAGGQAGTA
TH_3810|M.thermoresistible__bu --------------------------------------------------
MAB_4284c|M.abscessus_ATCC_199 VGGSTAVGGGAHTGVGGGVNTGVGVGSHVGGGISESLGGSGSVGGSTAVG
MAV_4827|M.avium_104 GGFGGGHAPAPGGGLGGGGGHAPAPGGGLGGFGGGHSGGGLGGLGGGHK-
MLBr_01556|M.leprae_Br4923 RIGDSVVAGDAYGRVRRMVDEHGVDIEAALPSSPVQVIGFTSVPGAGDNF
MSMEG_3202|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_3867|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb2648c|M.bovis_AF2122/97 --------------------------------------------------
Rv2615c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0806|M.marinum_M GTGGDATTKAAGGLDGARGNGGNGGNGGRGADGGAAIGSTAGGHGGDGGK
MUL_3318|M.ulcerans_Agy99 GQFGEP------------GTGGSGGNGGKGGDG-----------------
Mflv_5091|M.gilvum_PYR-GCK GKGGLFGGSGSAGKAG--ATGAAGASGGKGAQGATGAQGATG--------
TH_3810|M.thermoresistible__bu --------------------------------------------------
MAB_4284c|M.abscessus_ATCC_199 G-------------------------------------------------
MAV_4827|M.avium_104 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 LVVDEDRIARQIADRRSARKRNALAARSRKRISLEDLDSALKETSQLNLI
MSMEG_3202|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_3867|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb2648c|M.bovis_AF2122/97 --------------------------------------------------
Rv2615c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0806|M.marinum_M GTIGGNGGRGGNGGHGAQVVPAGARGGDGGSGGTGDVTGGAGGAGGNGGN
MUL_3318|M.ulcerans_Agy99 -----------------------AAGGNGGTGG----IGGTGGIGGDGRN
Mflv_5091|M.gilvum_PYR-GCK ---------------------AQGATGATGAAGKAGATGAAGAAGAAGSA
TH_3810|M.thermoresistible__bu --------------------------------------------------
MAB_4284c|M.abscessus_ATCC_199 ------------------------GAHTSAINDALGGSGSVGGSTAVGGG
MAV_4827|M.avium_104 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 LKGDNAGTVEALEEALMGIQVDDEVALRVIDRGVGGITETNVNLASASDA
MSMEG_3202|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_3867|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb2648c|M.bovis_AF2122/97 --------------------------------------------------
Rv2615c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0806|M.marinum_M GRNGGLGGAGGAGGNGGVTAASNIVTGNGGNGGNGGNGGPGNSSTGGNSG
MUL_3318|M.ulcerans_Agy99 GSTGAIGGNGGTGGTGG---------------------------------
Mflv_5091|M.gilvum_PYR-GCK GTTGAQGSAGTNGANGAN--------------------------------
TH_3810|M.thermoresistible__bu --------------------------------------------------
MAB_4284c|M.abscessus_ATCC_199 AHTSAINESMNSSINASHS-------------------------------
MAV_4827|M.avium_104 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 IIIGFNVRAEGKATELASR-------------------------------
MSMEG_3202|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_3867|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb2648c|M.bovis_AF2122/97 --------------------------------------------------
Rv2615c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0806|M.marinum_M NGGGGGLGGYGATGGTGGKGGNGGSGGIGGNGGNGGFGGGGTIRGDGGNG
MUL_3318|M.ulcerans_Agy99 ------------TGGTGGKSLGNLPSGDGGVGGNAGTGGNG---------
Mflv_5091|M.gilvum_PYR-GCK -----------GSQGAAGKAGATGTTGATGAAGEDGDAGSG---------
TH_3810|M.thermoresistible__bu --------------------------------------------------
MAB_4284c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_4827|M.avium_104 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 ------------EGVEIRYYLVIYQAIDDIEKALRGMLKPIYEENQLGRA
MSMEG_3202|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_3867|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb2648c|M.bovis_AF2122/97 --------------------------------------------------
Rv2615c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0806|M.marinum_M GNGGNGIGAGMDGGAGGKGGEGLTGGNGGNGGNGDNGVNGSNGGNGGKGG
MUL_3318|M.ulcerans_Agy99 -------GNATDGGAGGKGGDGAAGGTGGNAGEFQSLLGISKGGDGGTGG
Mflv_5091|M.gilvum_PYR-GCK -------GSGINGGAGGAGGAGGEGGEGGDGGKGGAGAEGGAGGGGGAGG
TH_3810|M.thermoresistible__bu --------------------------------------------------
MAB_4284c|M.abscessus_ATCC_199 --------SSMNESFGSSMGSHGAAGTGFEAQGHAQGHASETAHTTLPGA
MAV_4827|M.avium_104 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 EIRALFRSSKVGIIAGCIISSGVVRRNAKVRLLRDNIVVTDNLTVTSLRR
MSMEG_3202|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_3867|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb2648c|M.bovis_AF2122/97 --------------------------------------------------
Rv2615c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0806|M.marinum_M NAGNGTVVTGLGGNGGNGGDGGHGNGSNIAGGNGGSGGDGGTGATGGAGG
MUL_3318|M.ulcerans_Agy99 DGGTGTPVG-----------------------------------ADGLAG
Mflv_5091|M.gilvum_PYR-GCK SGAGG---------------------------------------AGGAGG
TH_3810|M.thermoresistible__bu --------------------------------------------------
MAB_4284c|M.abscessus_ATCC_199 EHH-----------------------------------------------
MAV_4827|M.avium_104 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 EKDD------------------------------------VTEVREGFEC
MSMEG_3202|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_3867|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb2648c|M.bovis_AF2122/97 --------------------------------------------------
Rv2615c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0806|M.marinum_M RGGAGGNGFNNGGDGGNGGNGGSGGNGVVRGNGGSGGNAGNGGNGVNGLF
MUL_3318|M.ulcerans_Agy99 TGGAGGFGIFPGGQTGQNGTDSKAG-------------------------
Mflv_5091|M.gilvum_PYR-GCK SGGSGG----SGGSGGSGGAAGDSDD------------------------
TH_3810|M.thermoresistible__bu --------------------------------------------------
MAB_4284c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_4827|M.avium_104 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 GMTLGYSDIKEGDVIESYELVEKQRA------------------------
MSMEG_3202|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_3867|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb2648c|M.bovis_AF2122/97 --------------------------------------------------
Rv2615c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0806|M.marinum_M TNRDGGDGGNGGNGGDGAIRGDGGAGGNGGNGVGTLNTVNPNGGDGGNGG
MUL_3318|M.ulcerans_Agy99 --------------------------------------------------
Mflv_5091|M.gilvum_PYR-GCK --------------------------------------------------
TH_3810|M.thermoresistible__bu --------------------------------------------------
MAB_4284c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_4827|M.avium_104 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 --------------------------------------------------
MSMEG_3202|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_3867|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb2648c|M.bovis_AF2122/97 --------------------------------------------------
Rv2615c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0806|M.marinum_M NGGNGNTGGTGGRGGNGGANGAVHGGDGGNGGKGGNGVIAGAGGDGGTGG
MUL_3318|M.ulcerans_Agy99 --------------------------------------------------
Mflv_5091|M.gilvum_PYR-GCK --------------------------------------------------
TH_3810|M.thermoresistible__bu --------------------------------------------------
MAB_4284c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_4827|M.avium_104 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 --------------------------------------------------
MSMEG_3202|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_3867|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb2648c|M.bovis_AF2122/97 --------------------------------------------------
Rv2615c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0806|M.marinum_M AGGGFQATGGSGGDGGAGGSGTTGGTGGRGGAGGAADGFRATGGAGGTGG
MUL_3318|M.ulcerans_Agy99 --------------------------------------------------
Mflv_5091|M.gilvum_PYR-GCK --------------------------------------------------
TH_3810|M.thermoresistible__bu --------------------------------------------------
MAB_4284c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_4827|M.avium_104 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 --------------------------------------------------
MSMEG_3202|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_3867|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb2648c|M.bovis_AF2122/97 --------------------------------------------------
Rv2615c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0806|M.marinum_M GGGNGTSTGGVGGNGGKGGGATDLESVAGSGGNGGNGGIGATGGNAGRGG
MUL_3318|M.ulcerans_Agy99 --------------------------------------------------
Mflv_5091|M.gilvum_PYR-GCK --------------------------------------------------
TH_3810|M.thermoresistible__bu --------------------------------------------------
MAB_4284c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_4827|M.avium_104 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 --------------------------------------------------
MSMEG_3202|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_3867|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb2648c|M.bovis_AF2122/97 --------------------------------------------------
Rv2615c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0806|M.marinum_M DGGSSTGFGGGGGMGGNGGTGGLGNAGYGGYGGDGGKGGDGGDTGGGVGR
MUL_3318|M.ulcerans_Agy99 --------------------------------------------------
Mflv_5091|M.gilvum_PYR-GCK --------------------------------------------------
TH_3810|M.thermoresistible__bu --------------------------------------------------
MAB_4284c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_4827|M.avium_104 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 --------------------------------------------------
MSMEG_3202|M.smegmatis_MC2_155 ----------------------------------------------
Mvan_3867|M.vanbaalenii_PYR-1 ----------------------------------------------
Mb2648c|M.bovis_AF2122/97 ----------------------------------------------
Rv2615c|M.tuberculosis_H37Rv ----------------------------------------------
MMAR_0806|M.marinum_M SGGTGGKGGASTLNGQQVAGGSGGSSKAPVGNRGSSGGTGGHPGGL
MUL_3318|M.ulcerans_Agy99 ----------------------------------------------
Mflv_5091|M.gilvum_PYR-GCK ----------------------------------------------
TH_3810|M.thermoresistible__bu ----------------------------------------------
MAB_4284c|M.abscessus_ATCC_199 ----------------------------------------------
MAV_4827|M.avium_104 ----------------------------------------------
MLBr_01556|M.leprae_Br4923 ----------------------------------------------