For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSGHNRERDAALPHGVQGAADPNFACTARAFSRLFPGRRFGGGALAVYQDGQPLVDIWTGYSDRAGTEYW TADTGAMVFSATKGLASTVIHRLADRGLIDYDTPVSAYWPEFAANGKAHITVREVMQHRAGLSQLNGVSK ADMLDHRVMEARIAEASVNRHLYGKSAYHALTYGWLMSGLVRAVTGQGMRELFRTELAEPLNTDGIHLGR PPVGAPTRAAQILGPQRTWPNHVFDFLAPRFAALEFSGFFGSTYCPGVMSLVQGDTPFLDAEIPAANGVA TARGLAKVYGAIANGGRFAGEEFLSAATVAGLSGRRSLSPDRNIGVPLAFHLGYHSVPFGVMAGFGHVGL GGSVGWADPASGLAVGFVHNRLLTPMVFDMGAFVGLHAVIRNDLARARRRGFEPVPDLGAAPYELSKPVA G
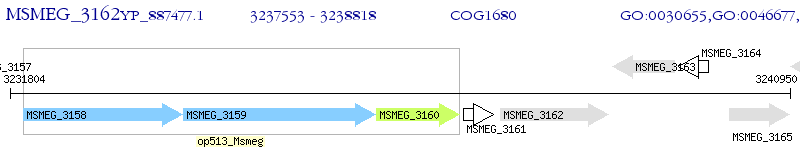
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3162 | - | - | 100% (421) | beta-lactamase |
| M. smegmatis MC2 155 | MSMEG_4680 | lipP | 2e-42 | 34.33% (367) | carboxylesterase, putative |
| M. smegmatis MC2 155 | MSMEG_3914 | - | 1e-37 | 32.01% (378) | beta-lactamase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1958 | lipD | 1e-143 | 59.36% (406) | lipase LIPD |
| M. gilvum PYR-GCK | Mflv_3647 | - | 1e-152 | 63.31% (417) | beta-lactamase |
| M. tuberculosis H37Rv | Rv1923 | lipD | 1e-143 | 59.36% (406) | lipase LIPD |
| M. leprae Br4923 | MLBr_00119 | lipE | 9e-27 | 29.90% (311) | putative hydrolase |
| M. abscessus ATCC 19977 | MAB_1386 | - | 1e-104 | 51.29% (388) | putative esterase/lipase/beta-lactamase |
| M. marinum M | MMAR_2305 | lipL | 1e-148 | 62.88% (423) | esterase LipL |
| M. avium 104 | MAV_3275 | - | 1e-150 | 64.15% (410) | beta-lactamase |
| M. thermoresistible (build 8) | TH_3778 | - | 1e-161 | 72.27% (375) | PUTATIVE beta-lactamase |
| M. ulcerans Agy99 | MUL_2951 | lipD | 1e-139 | 58.17% (404) | lipase LipD |
| M. vanbaalenii PYR-1 | Mvan_2765 | - | 1e-159 | 65.95% (417) | beta-lactamase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1958|M.bovis_AF2122/97 MDVAGLPRLAAGTQAAIIHGMAQPPSLLTTDNGL--PFGVQGACDSRFTG
Rv1923|M.tuberculosis_H37Rv MDVAGLPRLAAGTQAAIIHGMAQPPSLLTTDNGL--PFGVQGACDSRFTG
MUL_2951|M.ulcerans_Agy99 --------------------MAQP-SVPTTDDGL--PSGVAGACDSRFSG
MSMEG_3162|M.smegmatis_MC2_155 --------------MSGHNRERDAALPHG----------VQGAADPNFAC
TH_3778|M.thermoresistible__bu --------------------------------------------------
MMAR_2305|M.marinum_M -------------MTVDIGVNRRAVSSHGDPDAG--HR-VSGAADSQFAC
MAV_3275|M.avium_104 ---------------MDARVDRRAVRVHDDLDAG--HR-VSGAADSHFGC
Mflv_3647|M.gilvum_PYR-GCK -------------MTGVNSRERDEALSHGHGGHL--PRGVQGAADPNFTC
Mvan_2765|M.vanbaalenii_PYR-1 -------------MTGVNNPERDVALSHGHGAAL--PCGVQGAADPNFTL
MAB_1386|M.abscessus_ATCC_1997 ------MAIPLGPNVAATAPVRRDGLYPGDQPGLRLPNGVEGWARPELSA
MLBr_00119|M.leprae_Br4923 --------------MTSDGKIRVPTDLDAVTAIG--DEDHSDIDSAAVER
Mb1958|M.bovis_AF2122/97 VIRAFAGLYPGRKFGGGALSVYIDDRQVVDVWTGW--------SDRQGKV
Rv1923|M.tuberculosis_H37Rv VIRAFAGLYPGRKFGGGALSVYIDGRQVVDVWTGW--------SDRQGKV
MUL_2951|M.ulcerans_Agy99 VVRAFAGLYPGRKFGGGALSVYTEGRQVVDVWTGW--------ADRNGEV
MSMEG_3162|M.smegmatis_MC2_155 TARAFSRLFPGRRFGGGALAVYQDGQPLVDIWTGY--------SDRAGTE
TH_3778|M.thermoresistible__bu --------------------VYLHGEPVVDVWTGY--------ADRRGTE
MMAR_2305|M.marinum_M VVRMFSSMFPARRFGGGALAVYLDGQPVVDVWKGW--------ADREGEV
MAV_3275|M.avium_104 VVRSFASMFPARRFGGGALAVYVDGQPVVDVWTGW--------ADRAGQR
Mflv_3647|M.gilvum_PYR-GCK AVRAFAQMFPHRRFGGGALAVYLNGEPVVDVWTGW--------ADRRGKR
Mvan_2765|M.vanbaalenii_PYR-1 AVRAFAQMFPSRRLGGGALAVYLDGEPVVDVWTGW--------ADRRGTR
MAB_1386|M.abscessus_ATCC_1997 VVRTFEWMFTKYRLGGGGLSVYVDGEPALDIWAG---------AAAAGQG
MLBr_00119|M.leprae_Br4923 IWQATRHWYQAGMHPAIQLCIRQHGRVVLNRAIGHGWGNAPTDPHDAEKI
: ... :: * .
Mb1958|M.bovis_AF2122/97 PWTADTGAMVFSATKGLAATVIHRLVDRGLLSYDAPVAEYWPEFGANGKS
Rv1923|M.tuberculosis_H37Rv PWTADTGAMVFSATKGLAATVIHRLVDRGLLSYDAPVAEYWPEFGANGKS
MUL_2951|M.ulcerans_Agy99 PWTADTGAMVFSATKGLAATVIHRLVDRGLLDYDAPVADYWPEFGVKGKS
MSMEG_3162|M.smegmatis_MC2_155 YWTADTGAMVFSATKGLASTVIHRLADRGLIDYDTPVSAYWPEFAANGKA
TH_3778|M.thermoresistible__bu YWTADTGAMVFSATKGMAATVVHRLVDRGLLDYDTPVAEYWPEFGANGKA
MMAR_2305|M.marinum_M PWSADSAPMVFSATKGMAATVIHRLADRGLIDYEAPVAQYWPEFAANGKE
MAV_3275|M.avium_104 PWSENTAPMVFSATKGMAATVIHRLVDRGLIDYEAPVAEYWPAFAANGKA
Mflv_3647|M.gilvum_PYR-GCK RWTADTAPMVFSATKGVAATVIHRLADRGLIDYDAPVAEYWREFGVNGKS
Mvan_2765|M.vanbaalenii_PYR-1 RWTADTGPMVFSATKGVTATVIHRLHDRGLVDYDAPVAEYWREFGANGKS
MAB_1386|M.abscessus_ATCC_1997 -WTRDTGALVFSASKGIAATVIHRLVDRGLITYDAPVARYWPEFGANGKS
MLBr_00119|M.leprae_Br4923 PVTTDTPFCAYSAAKAITATVVHMLVERGHFSLDDRVCEYLPTYTSYGKH
: :: .:**:*.:::**:* * :** . : *. * : **
Mb1958|M.bovis_AF2122/97 EVTVSDVLRHRSGLAHLKGVD--KDEVMDHLLMEQKLAAAPLN-RQHGKL
Rv1923|M.tuberculosis_H37Rv EVTVSDVLRHRSGLAHLKGVD--KDEVMDHLLMEQKLAAAPLD-RQHGKL
MUL_2951|M.ulcerans_Agy99 AITVSDLLRHRSGLSHLKGVG--KKEILDHLRMEEKLAAAPLD-RTHGKM
MSMEG_3162|M.smegmatis_MC2_155 HITVREVMQHRAGLSQLNGVS--KADMLDHRVMEARIAEASVNRHLYGKS
TH_3778|M.thermoresistible__bu GITVRDVLRHRAGLSHLNGVS--KADLLDHRRMEERLAAAPVNRLQYGKP
MMAR_2305|M.marinum_M KLTVREVMRHAAGLSGLRGAS--QEDLLDHVVMEERLAAAAPG-RLLGKP
MAV_3275|M.avium_104 NMTVRQVMRHQAGLSGLRGAT--KEELLDHLVMEERLAAAAPG-RLLGKS
Mflv_3647|M.gilvum_PYR-GCK TITVRELMRHRAGLSQLNGAA--KTDLMDHLVMEERLAAAPAS-WLVGRP
Mvan_2765|M.vanbaalenii_PYR-1 AITVRDALRHRAGLSQLNGAG--KADLLDHAVMEQRLAAAPRS-WLYGKP
MAB_1386|M.abscessus_ATCC_1997 SITVRQVLDHRAGLSRLEGIAYHAEEIVDHELMERRLAAAPVD-KFYGKR
MLBr_00119|M.leprae_Br4923 RTTIRHVLTHSAGVPFPTGPRPDVTRADDHAYAQQKLSELRPFYRPGLVH
*: . : * :*:. * ** : :::
Mb1958|M.bovis_AF2122/97 AYHAVTYGWLLSGLARAVTGKGMRELFREELARPLNTDGIHLGRPPADSP
Rv1923|M.tuberculosis_H37Rv AYHAVTYGWLLSGLARAVTGKGMRELFREELARPLNTDGIHLGRPPADSP
MUL_2951|M.ulcerans_Agy99 AYHAVTYGWLLSGLARAVTGKGMRDLFREELARPLDTDGLHLGRPPGDAP
MSMEG_3162|M.smegmatis_MC2_155 AYHALTYGWLMSGLVRAVTGQGMRELFRTELAEPLNTDGIHLGRPPVGAP
TH_3778|M.thermoresistible__bu AYHALTYGWLLSGLVRAITGCGMRELVRTELAEPLNTDGLHLGRPPAGAP
MMAR_2305|M.marinum_M AYHALTFGWLMSGLARAVTGKDMRVLFREELAEPLDTDGFHLGRPPAGSP
MAV_3275|M.avium_104 AYHALTFGWLMSGLGRAVTGKGMRTLIREELAEPLGTDGLYLGRPPAGAP
Mflv_3647|M.gilvum_PYR-GCK AYHAFTFGWLLSGLARSVTGTGMRELIRTEVAAPLNTDGIHLGRPPVQAP
Mvan_2765|M.vanbaalenii_PYR-1 AYHALTFGWLLSGLARSITGKGMRELIRTEVAAPLNTDGLHLGRPPVQAP
MAB_1386|M.abscessus_ATCC_1997 AYHALTFGWLLAGLARSVTGKDMRELFRTEIADPLGVDGIHLGRPPRTAS
MLBr_00119|M.leprae_Br4923 IYHALTWGPLMREIIWTATGKEIRDILAAEILDPLGFRWTNFGVAEEDVP
***.*:* *: : : ** :* :. *: **. :* . .
Mb1958|M.bovis_AF2122/97 TKAAQTLLPQAKVPTP-----LLDFIAPKVAGL-SFSGLLGAVYFPGILS
Rv1923|M.tuberculosis_H37Rv TKAAQTLLPQAKVPTP-----LLDFIAPKVAGL-SFSGLLGAVYFPGILS
MUL_2951|M.ulcerans_Agy99 TTAAQTLLPQSKIPTP-----LLDFIAPKVAGL-SFSGLLGAVYFPGILS
MSMEG_3162|M.smegmatis_MC2_155 TRAAQILGPQRTWPNH-----VFDFLAPRFAAL-EFSGFFGSTYCPGVMS
TH_3778|M.thermoresistible__bu TKAAQILAPQGALDIP-----VVGAVVPRLAAS-QASAMFGAMYFPGMFS
MMAR_2305|M.marinum_M TQVAQIIMPQDIAANP-----LVTFAAHKIAN--QFSSGFRSMYFPGVIA
MAV_3275|M.avium_104 TRVAEIVAPQNLVGSS-----IVNYVSRKVAN--QLSGGFRAMYFPGMIA
Mflv_3647|M.gilvum_PYR-GCK TQPARIIGPQTRLQNP-----IFNAVAPHIAAL-PFSAGFGAMYFPGVKS
Mvan_2765|M.vanbaalenii_PYR-1 TEAAQIIGPQTRLQNP-----VFNLVAPRIAAL-PFSAGFGAIYLPGVKA
MAB_1386|M.abscessus_ATCC_1997 TKAASMYPFLDSMANKPAVGRLLPTVIRAIDRLPGFEGAIGTMYGPGMER
MLBr_00119|M.leprae_Br4923 LVALSHATGRP--------------LPPIIAAI--FRKAIGGTVH-EVIP
. : :
Mb1958|M.bovis_AF2122/97 LLQDD----MPFLDGEVPAVNGVVTARALAKTYGALANDGVIDGTRLLSS
Rv1923|M.tuberculosis_H37Rv LLQDD----MPFLDGEVPAVNGVVTARALAKTYGALANDGVIDGTRLLSS
MUL_2951|M.ulcerans_Agy99 MLQDD----MPFLDGEIPAVNGVVTARALAKTYAALANDGVIDGMRLLSS
MSMEG_3162|M.smegmatis_MC2_155 LVQGD----TPFLDAEIPAANGVATARGLAKVYGAIANGGRFAGEEFLSA
TH_3778|M.thermoresistible__bu VVQGD----IPFLDSEIPAANAVATARGLAKMYGAIANGGRIDDVRFLSP
MMAR_2305|M.marinum_M TVQGE----TPMLDAEIPAANGVVTARALARMYGAIANGGEIDGTQFLSR
MAV_3275|M.avium_104 AVQGD----TPLLDAEIPAANGVATARGLARMYGAIANGGEVDGIRFLSR
Mflv_3647|M.gilvum_PYR-GCK FVQGD----TPLLDGEIPAANGVATARALARMYGAIANEGRIDGLQYLSR
Mvan_2765|M.vanbaalenii_PYR-1 LVQGD----TPLLDGEIPAANGVATARALARMYGAIANGGRIDGTQYLSG
MAB_1386|M.abscessus_ATCC_1997 ILADDGTLNSALYDMQAPAANAVATAPALAKMYAALAGDGSVDGRELLSP
MLBr_00119|M.leprae_Br4923 YTNTP-----PFLTTIVPSSNTVSTANELSRFAEIWLRGGELDGIRVLRS
.: *: * * ** *:: * . . . *
Mb1958|M.bovis_AF2122/97 QAVRGLTGKSE-LWPDLNLGLP-----FTYHQGYQSS-PVPG-LLEGYGH
Rv1923|M.tuberculosis_H37Rv QAVRGLTGKSE-LWPDLNLGLP-----FTYHQGYQSS-PVPG-LLEGYGH
MUL_2951|M.ulcerans_Agy99 DAVSGLTGKAE-LWPDLNLGLP-----FTYHQGYQSP-PVPG-LLGGYGH
MSMEG_3162|M.smegmatis_MC2_155 ATVAGLSGRRS-LSPDRNIGVP-----LAFHLGYHSV-PFG--VMAGFGH
TH_3778|M.thermoresistible__bu EIVAGLAGRRN-LRPDRNLGVP-----MAFHLGYHGV-PFG--IMPGFGH
MMAR_2305|M.marinum_M ELVAGLTGERR-LQPDRNLFMP-----MAFHLGYHSL-PIGN-VMPGFGH
MAV_3275|M.avium_104 DLVAGLTGERS-LRPDRNLYVP-----LAFHLGYHSV-PIGG-VMPGFGH
Mflv_3647|M.gilvum_PYR-GCK ETTAVLAGRRS-LRLDHSMVMP-----LSFHLGYHGL-PIPG-LLPGFGH
Mvan_2765|M.vanbaalenii_PYR-1 ETAAALIGRPS-LRPDHSMVMP-----LSFHLGYHGL-PFPG-VLPGFGH
MAB_1386|M.abscessus_ATCC_1997 ETVAGLARKTN-LTIDRTIVFP-----MGMHLGYMSL-PMNG-FRGGFGH
MLBr_00119|M.leprae_Br4923 ETLLGAVAECRRLRPDFVVGLMPARWGTGFILGTNKFGPFGRNAPAAFGH
. * * : . * *. .:**
Mb1958|M.bovis_AF2122/97 IGLGGTIGWADPETGSAFGYVHNRLLT-LLLFDIGSFAGLAALLNSAVVA
Rv1923|M.tuberculosis_H37Rv IGLGGTIGWADPETGSAFGYVHNRLLT-LLLFDIGSFAGLAALLNSAVVA
MUL_2951|M.ulcerans_Agy99 IGLGGTIGWADPESGSAFGYVHNRLLT-LLLFDIGSFAGLAPLLSSAVST
MSMEG_3162|M.smegmatis_MC2_155 VGLGGSVGWADPASGLAVGFVHNRLLTPMVF-DMGAFVGLHAVIRNDLAR
TH_3778|M.thermoresistible__bu VGLGGSYGWAEPETGLAIGYVHNRLLTPMLI-DQAAFVGLNALIRRDAAA
MMAR_2305|M.marinum_M VGMGGSVGWTDPETGVAFALVHNRLLTPLVATDHAGFVAIYALIRQAAAK
MAV_3275|M.avium_104 VGMGGSLGWADPASGVAFSLVHNRLLTPLVMTDHAAFVGIYALIRRAAEQ
Mflv_3647|M.gilvum_PYR-GCK VGLGGSLGWADPETGLAFGFVHNRLLTPMVVSDQAGFVATAALLRRGASQ
Mvan_2765|M.vanbaalenii_PYR-1 VGLGGSLGWADPRSGLSFGYVHNRLLTPLVLSDQAGFVATAALIRRGAAA
MAB_1386|M.abscessus_ATCC_1997 IGLGGSMGWADPKRKIAVGFAHNRLPLTMAL-DQISFAFLWPQIVKAVG-
MLBr_00119|M.leprae_Br4923 LGLVNIAIWADPQRSMAAGLISS--GKPGQDPNAGRYGALMNTITAEIPV
:*: . *::* : . . : : :
Mb1958|M.bovis_AF2122/97 ARRDDPLEVPHFG-APYSEPRHEQAASGA--
Rv1923|M.tuberculosis_H37Rv ARRDDPLEVPHFG-APYSEPRHEQAASGA--
MUL_2951|M.ulcerans_Agy99 ARREGPLEVPHFG-ASYRETGQQPLAAGETS
MSMEG_3162|M.smegmatis_MC2_155 ARRRGFEPVPDLGAAPYELSK---PVAG---
TH_3778|M.thermoresistible__bu ARRRGYRPVGDLG-APFDMRK---RVAG---
MMAR_2305|M.marinum_M ARERGYQPVTGIG-APYSEPG---AVAG---
MAV_3275|M.avium_104 VRKHGFRPVTEFG-APFREPG---AVAG---
Mflv_3647|M.gilvum_PYR-GCK ARKNGYRRVREYG-SPFSDVA---ATAV---
Mvan_2765|M.vanbaalenii_PYR-1 ARRNGYRPVREYG-APFTDLT---ATAV---
MAB_1386|M.abscessus_ATCC_1997 -------------------------------
MLBr_00119|M.leprae_Br4923 H------------------------------