For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TDLIEETPSTSTATGSGRTRRQNIIVRFNLLFIFIALFAVASVTAPQFLTSQNLANLLQQSSLTGIVAIG MTVVILTSGIDLSVGSVAALSGMLVAILIGMGIAWPLAMVVAIMSGLVLGGIMGGLSAYLALPAFMVTLA GMQSIRGLTFLTTNGTPTTAEIPIGLRFLGAGSVAGIPVVGLIFVLTTVIVGLVLRRTTFGEHVYAVGSN AKAARLSGLPVQRITTAAYVICSGLAALTGVLLTARLTIGQPTANTGLELDAIAAVVLGGTSLFGGKGGV MGTFIAVLLLSVLRNLFNLLGLSSFFQMLVTGLILVAALIMNYVLDRQAKTS*
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
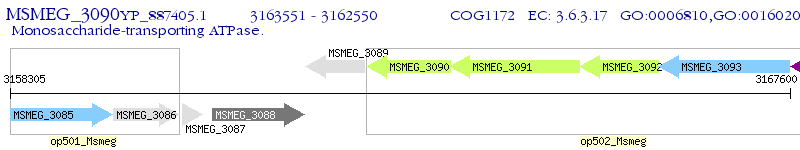
| M. smegmatis MC2 155 | MSMEG_3090 | - | - | 100% (333) | ribose transport system permease protein RbsC |
| M. smegmatis MC2 155 | MSMEG_4171 | - | 4e-60 | 38.60% (329) | ribose transport system permease protein RbsC |
| M. smegmatis MC2 155 | MSMEG_4001 | - | 7e-50 | 36.20% (326) | ribose transport system permease protein RbsC |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2606 | - | 3e-21 | 25.31% (320) | inner-membrane translocator |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_00397 | - | 2e-46 | 35.62% (292) | putative transporter protein |
| M. abscessus ATCC 19977 | MAB_2624c | - | 7e-10 | 25.93% (297) | high-affinity branched-chain amino acid ABC transporter (LivM) |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_2329 | - | 2e-11 | 26.37% (292) | branched-chain amino acid ABC-type transport system, permease |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_5291 | - | 9e-41 | 30.29% (373) | inner-membrane translocator |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_3090|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00397|M.leprae_Br4923 MDHTTKCDAEQYFQAIVTSMADGVIVVDIDGRIESINPAATRILGLRAHD
Mvan_5291|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2606|M.gilvum_PYR-GCK --------------------------------------------------
TH_2329|M.thermoresistible__bu --------------------------------------------------
MAB_2624c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_3090|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00397|M.leprae_Br4923 VVDMKHGHPFCFYDTDNQRVDLEREVMRVVRREVTTVSKVVGIDQHSGQR
Mvan_5291|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2606|M.gilvum_PYR-GCK --------------------------------------------------
TH_2329|M.thermoresistible__bu --------------------------------------------------
MAB_2624c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_3090|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00397|M.leprae_Br4923 LWLSVNVSLLAYKAPPHSALVVSFSDISAHHLSIERLTYEATHDCLTGLA
Mvan_5291|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2606|M.gilvum_PYR-GCK --------------------------------------------------
TH_2329|M.thermoresistible__bu --------------------------------------------------
MAB_2624c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_3090|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00397|M.leprae_Br4923 NRRFAEDQITKSLQHDERSRLAAVLLLDLDDFKVINDSLGHDVGDAVLQT
Mvan_5291|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2606|M.gilvum_PYR-GCK --------------------------------------------------
TH_2329|M.thermoresistible__bu --------------------------------------------------
MAB_2624c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_3090|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00397|M.leprae_Br4923 VAQRLRSAVRPDDVVARLGGDEFIVLLRGPLSDMNANDVAKRLHTTLSES
Mvan_5291|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2606|M.gilvum_PYR-GCK --------------------------------------------------
TH_2329|M.thermoresistible__bu --------------------------------------------------
MAB_2624c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_3090|M.smegmatis_MC2_155 --------------------------MTDLIEETPSTSTATGSGRTRRQN
MLBr_00397|M.leprae_Br4923 LVVDQLTVPIGASVGILEVRPDDRRRAADILRDADSAMYAAKNKKQCAVT
Mvan_5291|M.vanbaalenii_PYR-1 ---------MTTVRSDSDVSLADADFSGDTRADETFGAAVRGYLQRVRGG
Mflv_2606|M.gilvum_PYR-GCK --------------------MSTKEALDVGGHKVVRDERVKERNRLQRLL
TH_2329|M.thermoresistible__bu -------------------------------MDILIGQLATG-LSLGSIL
MAB_2624c|M.abscessus_ATCC_199 ---------------------------MSAARALAPGDSIRRWWSELARP
MSMEG_3090|M.smegmatis_MC2_155 IIVRFNLLFIFIALFAVASVTAPQFLTSQNLANLLQQSSLTGIVAIGMTV
MLBr_00397|M.leprae_Br4923 PQQLVPFVALIALFVFFTAAAGAKFYAPSNLLVILQQTVVLAIVGYGMTF
Mvan_5291|M.vanbaalenii_PYR-1 DMGSLPAVLGLVVLFVVFGLANDRFLSALNLANLITQAGSICVLAMGLVF
Mflv_2606|M.gilvum_PYR-GCK IRPEMGAGIGAIGIFVFFLVVAPPFREASSLATVLYASSTIGIMACGVAV
TH_2329|M.thermoresistible__bu LLAALGLALTFGQMGVINMAHGEFIMVGCYTAYVVQQ--IVSDAGVSLLV
MAB_2624c|M.abscessus_ATCC_199 VQWVYGVVGFALLALLPVWKIPYLDTPNTSFGGVMAQFAMVALIAIGLNV
:: . .: .
MSMEG_3090|M.smegmatis_MC2_155 VI-LTSGIDLSVGSVAALSGMLVAILIGMG---------------IAWPL
MLBr_00397|M.leprae_Br4923 VI-MAGSVDLSVGSIVALTGVTAALVAAQ-----------------NQFA
Mvan_5291|M.vanbaalenii_PYR-1 VL-LLGDIDLSAGVAGGVSACAMALIVVNLS--------------WPWWA
Mflv_2606|M.gilvum_PYR-GCK LM-IGGEFDLSAGVAVTFSSLAASMLAYNLH--------------LNLWV
TH_2329|M.thermoresistible__bu SL-FVG---------FLVGGAMGALLEVTLI--------------RRMYH
MAB_2624c|M.abscessus_ATCC_199 VVGQAGLLDLGYVGFYAIGAYTVALLTSPDSPWNRMGTSAFFSSDWAWLS
: . . . :::
MSMEG_3090|M.smegmatis_MC2_155 AMVVAIMSGLVLGGIMGGLSAYLALPAFMVTLAGMQSIRGLTFLTTNGT-
MLBr_00397|M.leprae_Br4923 AIVTALLVGLAAGMVNGIVFAYGKIPSFVSTLGMLQVCRGITLMISDSS-
Mvan_5291|M.vanbaalenii_PYR-1 ALLVGIACGALIGLAIGLLRAKLGIPSFVVTLAFFLGLQGVTLKLIGEGG
Mflv_2606|M.gilvum_PYR-GCK GALLALVLALTVGFFNGFLVMKTKIPSFLITLSTFFMLAGINLAVTKLVA
TH_2329|M.thermoresistible__bu RPLDTLLVTFGVGLILQQVARDIFGAPAVNVLAPAWLGGGVEIFG-----
MAB_2624c|M.abscessus_ATCC_199 CLPIAVAITACFGLILGTPTLRLRGDYLAIVTLGFGEIIRLLADNLDVTN
: * . :
MSMEG_3090|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00397|M.leprae_Br4923 --------------------------------------------------
Mvan_5291|M.vanbaalenii_PYR-1 SVRVDDPVIRGLTISNLPVAAGWAIAGVVVLGFAGLELHQHRRKIALRLS
Mflv_2606|M.gilvum_PYR-GCK G-------------------------------------------------
TH_2329|M.thermoresistible__bu --------------------------------------------------
MAB_2624c|M.abscessus_ATCC_199 GPR-----------------------------------------------
MSMEG_3090|M.smegmatis_MC2_155 -------------------PTTAEIPIGLRFLGAGSVA--------GIPV
MLBr_00397|M.leprae_Br4923 -------------------AKPMPFHG-----ILGAMG--------AMPW
Mvan_5291|M.vanbaalenii_PYR-1 HSPLGVVVARVGAVAAVVLGVTYVMSVNRSVSAVADIR--------GIPY
Mflv_2606|M.gilvum_PYR-GCK -----------------QVATQSVSDMQGWDSAQKVFASSFTLFGVGIRI
TH_2329|M.thermoresistible__bu -------------------------------AVVPKTR----------LF
MAB_2624c|M.abscessus_ATCC_199 ------------GLHKVAYPHVGEQHVHGGGVFSSANSGGHLNYGVWWYW
MSMEG_3090|M.smegmatis_MC2_155 VGLIFVLTTVIVGLVLRRTTFGEHVYAVGSNAKAARLSGLPVQRITTAAY
MLBr_00397|M.leprae_Br4923 ILIVCLFVTILAGILFQFTMFGRWVKAIGGNERVATLAGVPTRGIKVAIF
Mvan_5291|M.vanbaalenii_PYR-1 VLPLMLVLLIAMTVVLRRTSYGRHIYAVGGNAEAARRAGISVDRIRVSVF
Mflv_2606|M.gilvum_PYR-GCK TVVWWLVFTAVATWVLFKTKVGNWIFAVGGDADSARAVGVPVTKVKIGLF
TH_2329|M.thermoresistible__bu ILVLAVVCVAALAAVLKYSPAGRRIRAVVQNRDLAETSGISSRTTDVTTF
MAB_2624c|M.abscessus_ATCC_199 LGLCLIAAVLVLMGNLERSRVGRSWVAIREDEDAAEMMGVPTFRFKLWAF
: : : *. *: : * *:. :
MSMEG_3090|M.smegmatis_MC2_155 VICSGLAALTGVLLTARLTIGQPTANTG-LELDAIAAVVLGGTSLFGGKG
MLBr_00397|M.leprae_Br4923 AICGLTAGLGGIVLASRLGAGTPTAATG-FEIDVIAAVVIGGTPLTGGLG
Mvan_5291|M.vanbaalenii_PYR-1 VVCSSLAALSGIIAASYAGKVSASSGAGNTLLYAVGAAVIGGTSLFGGKG
Mflv_2606|M.gilvum_PYR-GCK MFVGFCAWFVGMHLLFAFNTVQSGQGIG-NEFFYIIAAVIGGCLLTGGYG
TH_2329|M.thermoresistible__bu FIGSGLAAVAGVALT-LIGSTSPTIGQS-YLIDAFLVVVVG------GLG
MAB_2624c|M.abscessus_ATCC_199 VIGASIGGLSGALYAGQVQYVASPTFNIINSMLFLCAVVLG------GQG
. . . . * . : . ..*:* * *
MSMEG_3090|M.smegmatis_MC2_155 GVMGTFIAVLLLSVLRN-----LFNLLGLSSFFQMLVTG-LILVAALIMN
MLBr_00397|M.leprae_Br4923 RLSGTLIGAIIISMLSN-----GMVFMGVGNAASQIIKG-IMLAAAVFVF
Mvan_5291|M.vanbaalenii_PYR-1 RAIDAVIGGVVVATIAN-----GLGLLNQSSYINFLVTGGVLLLAASVDA
Mflv_2606|M.gilvum_PYR-GCK TAIGAAIGALIFGMTNQ-----GIVYAGWDPDWFKFFLGAMLLFAVIANN
TH_2329|M.thermoresistible__bu QIKGTVIAALGLGFLNS-----FIEYSTTASLAKVIVFALIVIFLQVRPQ
MAB_2624c|M.abscessus_ATCC_199 NKLGVVVGAFIIVYLPNRLLGVQVQGIDLGNLKYLFFGLALVVLMLFRPQ
.. :. . . . . :. :::
MSMEG_3090|M.smegmatis_MC2_155 YVLDRQAKTS----------------------
MLBr_00397|M.leprae_Br4923 LQRRKIGIIK----------------------
Mvan_5291|M.vanbaalenii_PYR-1 ISRRRRSSTGLG--------------------
Mflv_2606|M.gilvum_PYR-GCK AFRSYAARK-----------------------
TH_2329|M.thermoresistible__bu GLFTVRTRSLA---------------------
MAB_2624c|M.abscessus_ATCC_199 GLFPAHQKLMAYWRKTSERLSAQKPDEVEAGA