For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTSDVTQRTFYCGLDAALAIVGGRWKFLIVWQLAVQGAQRYGQLRARVEGVSEKVLIGALRDLEADGVVE RRDHRTVPPHVEYSLTPSGVELAKVLEPLCRWGYDHMKVHTEARTEQMAHRAELAALA
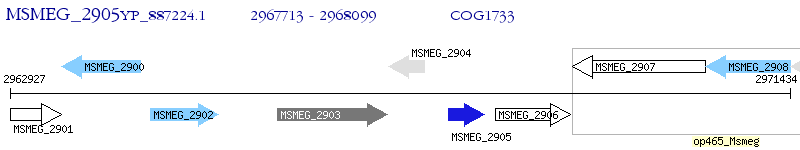
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2905 | - | - | 100% (128) | transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_6344 | - | 6e-23 | 45.28% (106) | transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_2849 | - | 5e-15 | 35.92% (103) | transcriptional regulatory protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3122 | - | 9e-09 | 34.29% (105) | putative transcriptional regulatory protein |
| M. gilvum PYR-GCK | Mflv_5238 | - | 2e-16 | 41.94% (124) | HxlR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv3095 | - | 1e-08 | 34.29% (105) | putative transcriptional regulatory protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3696c | - | 3e-13 | 40.66% (91) | putative transcriptional regulator |
| M. marinum M | MMAR_1626 | - | 6e-09 | 35.48% (93) | transcriptional regulator |
| M. avium 104 | MAV_4183 | - | 2e-05 | 27.55% (98) | putative transcriptional regulator family protein |
| M. thermoresistible (build 8) | TH_2405 | - | 2e-09 | 33.66% (101) | putative transcriptional regulator |
| M. ulcerans Agy99 | MUL_3475 | - | 7e-09 | 35.24% (105) | transcriptional regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_1032 | - | 1e-16 | 41.09% (129) | HxlR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_5238|M.gilvum_PYR-GCK ----MGRLADVSRRTYGQYCGLAHALDVVGERWTLLIVRELA-SGPKRYT
Mvan_1032|M.vanbaalenii_PYR-1 ----MSTIVGMTRRSYGQFCGLAHALDVVGERWTLLIVRELG-SGPKRYG
MSMEG_2905|M.smegmatis_MC2_155 ------MTSDVTQRTF--YCGLDAALAIVGGRWKFLIVWQLAVQGAQRYG
Mb3122|M.bovis_AF2122/97 -----MAVSDLSHRFEG--ESVGRALELVGERWTLLILREAF-FGVRRFG
Rv3095|M.tuberculosis_H37Rv -----MAVSDLSHRFEG--ESVGRALELVGERWTLLILREAF-FGVRRFG
MUL_3475|M.ulcerans_Agy99 -----MGAADLSHRFEG--ESVGRALELVGERWTLLILREAF-FGVQRFG
TH_2405|M.thermoresistible__bu ----------MASRTYGQHCALAKSLDLVGDRWTLLIVRELL-DGPRRYS
MMAR_1626|M.marinum_M ------------MSTYSQFCPVAKAMELLDERWTLLVVRELL-LGSTHFN
MAB_3696c|M.abscessus_ATCC_199 MYPSSQPTDDWWGNLFDTQCPTRIVLDRIGDKWTVLVVAALA-DGPMRFT
MAV_4183|M.avium_104 MTLLQGPLADRDAWSAVGECAIEKTMAVVGTKSAMLIMREAY-YGTTRFD
: :. : .*:: * ::
Mflv_5238|M.gilvum_PYR-GCK ELADALAGIGTSLLATRVRQLETDGVVQRRLALDQPG-STVVYELSDAGR
Mvan_1032|M.vanbaalenii_PYR-1 ELADALAGIGTSLLATRMRQLETDGVIQRRLDLDQPG-SAVVYELSDAGR
MSMEG_2905|M.smegmatis_MC2_155 QLRARVEGVSEKVLIGALRDLEADGVVERRDHRTVP--PHVEYSLTPSGV
Mb3122|M.bovis_AF2122/97 QLARNLG-IPRPTLSSRLRMLVEVGLFDRVPYSSDP--ERHEYRLTEAGR
Rv3095|M.tuberculosis_H37Rv QLARNLG-IPRPTLSSRLRMLVEVGLFDRVPYSSDP--ERHEYRLTEAGR
MUL_3475|M.ulcerans_Agy99 QLARNLN-IPRPTLSSRLRMLVNAGLLDRVPYSQDP--ERHEYRLTDTGH
TH_2405|M.thermoresistible__bu DLLTALRPIATDMLATRLRHLEEHHLVRRR-ELPKPA-SGTVYELTADGA
MMAR_1626|M.marinum_M DLRRGVPKMSPALLSKRLKSLTKAGVVERSDIEGRT-----SYSLTTCGR
MAB_3696c|M.abscessus_ATCC_199 ALRDRIGGVTGKVLTSTLRSMLRDGLVKRTAYATVP--PKVEYQLTELGL
MAV_4183|M.avium_104 DFARRVG-ITKAATSARLAELVELGLLKRRPYREPGQRSRDEYVLTQAGI
: : : : : :. * * *: *
Mflv_5238|M.gilvum_PYR-GCK ELAAAVVPLAMWGARHQMAD-ADVCHEAFRAEWLLSFLAADVRRDGPHDL
Mvan_1032|M.vanbaalenii_PYR-1 ELAEAMIPLAMWGARHQMSH-ADPCAEAFRAEWLLAFLAADLRGGIPEGL
MSMEG_2905|M.smegmatis_MC2_155 ELAKVLEPLCRWGYDHMKVH-TEARTEQMAHRAELAALA-----------
Mb3122|M.bovis_AF2122/97 DLFAAIVVLMQWGDEYLPRP-EGPPIKLRHHTCGEHADPRLICTHCGEEI
Rv3095|M.tuberculosis_H37Rv DLFAAIVVLMQWGDEYLPRP-EGPPIKLRHHTCGEHADPRLICTHCGEEI
MUL_3475|M.ulcerans_Agy99 ELFAAIVVLMQWGDKHLPNP-DGPPIVLRHH-CGQFVEPRLICAHCGEEI
TH_2405|M.thermoresistible__bu ALEAVVHAFIRWG-RSLLET-RRAGDVVRPHWLARAVRAHVRPDRTGPPL
MMAR_1626|M.marinum_M ELATVVDALGEWGIRWIPELGDEDLDPHLLMWDIRRTVPIDAWPRARTVL
MAB_3696c|M.abscessus_ATCC_199 SLREPIDQLRTWAERNVAHIVRNREAHDGDMPSTDTAIG-----------
MAV_4183|M.avium_104 DFMPVVWAMFEWGRRHLPGR---HRLQLTHLGCGAEVGVEIRCAEG----
: : : *.
Mflv_5238|M.gilvum_PYR-GCK DAVYEFHIDDSTACLRLRDGGVTVTPGAPATPGDVVVRSRASTIAEIVGH
Mvan_1032|M.vanbaalenii_PYR-1 ASVYEFHVDDSTACLQIRDGRVSVRPGPCATPADVTVRATARAIAAVVGQ
MSMEG_2905|M.smegmatis_MC2_155 --------------------------------------------------
Mb3122|M.bovis_AF2122/97 TAR-------------------NVTPEPGPGFKAKLASS-----------
Rv3095|M.tuberculosis_H37Rv TAR-------------------NVTPEPGPGFKAKLASS-----------
MUL_3475|M.ulcerans_Agy99 TAR-------------------NVTPEAGPGYGDAVAAPN----------
TH_2405|M.thermoresistible__bu SVRLVTPEGAAAVRIS-----ADAVEDLDDDTAVDVTLTGAAEVLAAALH
MMAR_1626|M.marinum_M AFRLAGVAPKAARWWLLVSEGEADVCDFDPGYQVAATVETSLLTLTQLWR
MAB_3696c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_4183|M.avium_104 ----------------------HLVPPDELGMRLAKKSG-----------
Mflv_5238|M.gilvum_PYR-GCK KIAVAD-ALADGSLEITGDADKISALVGVIEKRLAR------
Mvan_1032|M.vanbaalenii_PYR-1 KDRLED-AVAEGRIEVTGDPAAIATLIGAVEKRLA-------
MSMEG_2905|M.smegmatis_MC2_155 ------------------------------------------
Mb3122|M.bovis_AF2122/97 ------------------------------------------
Rv3095|M.tuberculosis_H37Rv ------------------------------------------
MUL_3475|M.ulcerans_Agy99 ------------------------------------------
TH_2405|M.thermoresistible__bu PDRAAE-LVAQGRLRVDGRADALRRLAEVVTAPAGRAVPDRS
MMAR_1626|M.marinum_M GDQSWSRAMLNGGVRIDGPAQVRRSVPAWLGQSRLAAVPRPA
MAB_3696c|M.abscessus_ATCC_199 ------------------------------------------
MAV_4183|M.avium_104 ------------------------------------------