For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MGRLADVSRRTYGQYCGLAHALDVVGERWTLLIVRELASGPKRYTELADALAGIGTSLLATRVRQLETDG VVQRRLALDQPGSTVVYELSDAGRELAAAVVPLAMWGARHQMADADVCHEAFRAEWLLSFLAADVRRDGP HDLDAVYEFHIDDSTACLRLRDGGVTVTPGAPATPGDVVVRSRASTIAEIVGHKIAVADALADGSLEITG DADKISALVGVIEKRLAR
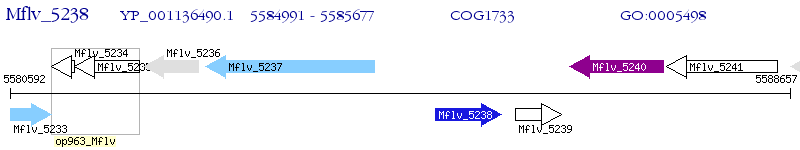
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_5238 | - | - | 100% (228) | HxlR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_2233 | - | 2e-27 | 36.06% (208) | HxlR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_3781 | - | 9e-06 | 28.83% (111) | HxlR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1754c | - | 8e-20 | 31.68% (202) | hypothetical protein Mb1754c |
| M. tuberculosis H37Rv | Rv1725c | - | 8e-20 | 31.68% (202) | hypothetical protein Rv1725c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3456 | - | 1e-11 | 38.04% (92) | HxlR family transcriptional regulator |
| M. marinum M | MMAR_1147 | - | 6e-20 | 36.41% (217) | transcriptional regulatory protein |
| M. avium 104 | MAV_4183 | - | 1e-05 | 30.63% (111) | putative transcriptional regulator family protein |
| M. smegmatis MC2 155 | MSMEG_5032 | - | 3e-28 | 37.22% (223) | putative transcriptional regulator |
| M. thermoresistible (build 8) | TH_2405 | - | 4e-28 | 37.16% (218) | putative transcriptional regulator |
| M. ulcerans Agy99 | MUL_0912 | - | 3e-20 | 35.91% (220) | transcriptional regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_1032 | - | 3e-91 | 71.37% (227) | HxlR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1754c|M.bovis_AF2122/97 ------------MQPYGQYCPVARAAELLGDRWTLLIVRELLFGPLRFTE
Rv1725c|M.tuberculosis_H37Rv ------------MQPYGQYCPVARAAELLGDRWTLLIVRELLFGPLRFTE
Mflv_5238|M.gilvum_PYR-GCK ----MGRLADVSRRTYGQYCGLAHALDVVGERWTLLIVRELASGPKRYTE
Mvan_1032|M.vanbaalenii_PYR-1 ----MSTIVGMTRRSYGQFCGLAHALDVVGERWTLLIVRELGSGPKRYGE
MSMEG_5032|M.smegmatis_MC2_155 ----------MASRSYGQYCALAKSLDLVGDRWTLLVVRELLDGPRRYGD
TH_2405|M.thermoresistible__bu ----------MASRTYGQHCALAKSLDLVGDRWTLLIVRELLDGPRRYSD
MMAR_1147|M.marinum_M ----------MSARNYNQNCPIARGLDILGERWTLLILRELVGGPRRYGD
MUL_0912|M.ulcerans_Agy99 ----------MSARNYNQNCPIARGLDILGERWTLLILRELVGGPRRYGD
MAB_3456|M.abscessus_ATCC_1997 -----------MDFTTMRNCSISNALDVIGERWTLVALREIMLGNRRFET
MAV_4183|M.avium_104 MTLLQGPLADRDAWSAVGECAIEKTMAVVGTKSAMLIMREAYYGTTRFDD
* : . ::* : ::: :** * *:
Mb1754c|M.bovis_AF2122/97 IERGLPGISRSVLAQRLRRLQHDRIIEAV-PEHTGGG----YRFTVAGEE
Rv1725c|M.tuberculosis_H37Rv IERGLPGISRSVLAQRLRRLQHDRIIEAV-PEHTGGG----YRFTVAGEE
Mflv_5238|M.gilvum_PYR-GCK LADALAGIGTSLLATRVRQLETDGVVQRR-LALDQPGSTVVYELSDAGRE
Mvan_1032|M.vanbaalenii_PYR-1 LADALAGIGTSLLATRMRQLETDGVIQRR-LDLDQPGSAVVYELSDAGRE
MSMEG_5032|M.smegmatis_MC2_155 LLSALSPIATDVLAGRLRDLEANNLVHK--RAMPRPSATTVYALTADG-R
TH_2405|M.thermoresistible__bu LLTALRPIATDMLATRLRHLEEHHLVRR--RELPKPASGTVYELTADG-A
MMAR_1147|M.marinum_M LRAELAGIATNLLAQRLAELQERGLVDR--TELPPPIGRTVYTLSDGGWR
MUL_0912|M.ulcerans_Agy99 LRAELAGIATNLLAQRLAELQERGLVDR--TELPPPIGRTVYTLSDGGWR
MAB_3456|M.abscessus_ATCC_1997 IVRNTG-ASRDILATRLRKLVDAGVLEKR-RYEDRPP-RYEYLLTQSGQA
MAV_4183|M.avium_104 FARRVG-ITKAATSARLAELVELGLLKRRPYREPGQRSRDEYVLTQAGID
: : *: * :: * :: *
Mb1754c|M.bovis_AF2122/97 LRPVLQTLGDWVSRWLMA--DPTPAECDPELLTLWISRRVNTEALPGRRV
Rv1725c|M.tuberculosis_H37Rv LRPVLQTLGDWVSRWLMA--DPTPAECDPELLTLWISRRVNTEALPGRRV
Mflv_5238|M.gilvum_PYR-GCK LAAAVVPLAMWGARHQMADADVCHEAFRAEWLLSFLAADVRRDGPHDLDA
Mvan_1032|M.vanbaalenii_PYR-1 LAEAMIPLAMWGARHQMSHADPCAEAFRAEWLLAFLAADLRGGIPEGLAS
MSMEG_5032|M.smegmatis_MC2_155 ALEDVINAFARWGRHLLANRDRKETVRPE--WLVRAVRAYVRDDRTGPPV
TH_2405|M.thermoresistible__bu ALEAVVHAFIRWGRSLLETRRAGDVVRPH--WLARAVRAHVRPDRTGPPL
MMAR_1147|M.marinum_M RVLPVLQTLAWFGLDLIDPIDDSSVVSPLNGFLAGILLGFDPARAPGLAA
MUL_0912|M.ulcerans_Agy99 RVLPVLQTLAWFGLDLIDPIDDSSVVSPLNGFLAGILLGFDPARAPGLAA
MAB_3456|M.abscessus_ATCC_1997 LRPVLFALMEWGDKFVTK----------------------------GPPP
MAV_4183|M.avium_104 FMPVVWAMFEWGRRHLPG------------------------------RH
:
Mb1754c|M.bovis_AF2122/97 VVEFRYHGERPLWAWLVLEPGDISVCLHDPCLPVDLTVRGHPRDLYRVYS
Rv1725c|M.tuberculosis_H37Rv VVEFRYHGERPLWAWLVLEPGDISVCLHDPCLPVDLTVRGHPRDLYRVYS
Mflv_5238|M.gilvum_PYR-GCK VYEFHIDDST---ACLRLRDGGVTVTPGAPATPGDVVVRSRASTIAEIVG
Mvan_1032|M.vanbaalenii_PYR-1 VYEFHVDDST---ACLQIRDGRVSVRPGPCATPADVTVRATARAIAAVVG
MSMEG_5032|M.smegmatis_MC2_155 TVRMVTPEGQ---AAVEINEH--TVESLDDEADVDVTLTGAAEVLAAAMD
TH_2405|M.thermoresistible__bu SVRLVTPEGA---AAVRISAD--AVEDLDDDTAVDVTLTGAAEVLAAALH
MMAR_1147|M.marinum_M TYRAEIDGRR---FEFVVERG--RLGPARGAPTVTISAA-ALDLVTARLG
MUL_0912|M.ulcerans_Agy99 TYRAEIDGRR---FEFAVERG--RLGPARGAPTVTISVA-ALDLVTARLG
MAB_3456|M.abscessus_ATCC_1997 SIWEHQCGSV-------------------------LHIQPTCESCGEAVT
MAV_4183|M.avium_104 RLQLTHLGCG---------------------AEVGVEIRCAEGHLVPPDE
:
Mb1754c|M.bovis_AF2122/97 GRSTLAAEISAERIELDGLPAMRRAFPSWMAWSPFAPAMRQAVVSVDQMP
Rv1725c|M.tuberculosis_H37Rv GRSTLAAEISAERIELDGLPAMRRAFPSWMAWSPFAPAMRQAVVSVDQMP
Mflv_5238|M.gilvum_PYR-GCK HKIAVADALADGSLEITGDADKISALVGVIEK-------RLAR-------
Mvan_1032|M.vanbaalenii_PYR-1 QKDRLEDAVAEGRIEVTGDPAAIATLIGAVEK-------RLA--------
MSMEG_5032|M.smegmatis_MC2_155 P-GQVENLVAAGLLTIDGDARAVRRLAKAFAP----PRVRV---------
TH_2405|M.thermoresistible__bu P-DRAAELVAQGRLRVDGRADALRRLAEVVTA----PAGRAVPDRS----
MMAR_1147|M.marinum_M A-DEATRKAALGRIDFDGDPGAIDALCTAFSLSGNSPRARGADGRS----
MUL_0912|M.ulcerans_Agy99 A-DEATRKAALGRIDFDGDPGAIDALCTAFSLSGNSPRARGADGRS----
MAB_3456|M.abscessus_ATCC_1997 FDDLTPRRLGRVR-------------------------------------
MAV_4183|M.avium_104 LGMRLAKKSG----------------------------------------
.
Mb1754c|M.bovis_AF2122/97 EAHGG
Rv1725c|M.tuberculosis_H37Rv EAHGG
Mflv_5238|M.gilvum_PYR-GCK -----
Mvan_1032|M.vanbaalenii_PYR-1 -----
MSMEG_5032|M.smegmatis_MC2_155 -----
TH_2405|M.thermoresistible__bu -----
MMAR_1147|M.marinum_M -----
MUL_0912|M.ulcerans_Agy99 -----
MAB_3456|M.abscessus_ATCC_1997 -----
MAV_4183|M.avium_104 -----