For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VCGYRDAVRPYARLRLCRRREELSVATVVVADDHSLIREGVVRVLERGGHRVLAVAADAVELLVQVDIHR PDVVVTDIEMPPSVARLKFVYECGAGFRRRSWSTRMGGRVGSGPR
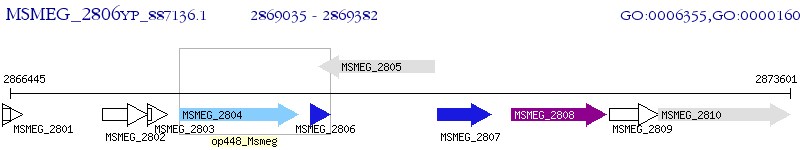
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2806 | - | - | 100% (115) | two-component system response regulator |
| M. smegmatis MC2 155 | MSMEG_0459 | - | 2e-13 | 57.14% (56) | two-component system response regulator |
| M. smegmatis MC2 155 | MSMEG_4969 | - | 2e-10 | 44.64% (56) | two-component system response regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1652 | - | 3e-07 | 33.33% (54) | two-component system transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_0968 | - | 5e-11 | 51.79% (56) | two component LuxR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv1626 | - | 3e-07 | 33.33% (54) | two-component system transcriptional regulator |
| M. leprae Br4923 | MLBr_01286 | - | 7e-07 | 35.19% (54) | putative two-component response regulatory protein |
| M. abscessus ATCC 19977 | MAB_4498c | - | 1e-10 | 49.18% (61) | two-component LuxR family response regulator |
| M. marinum M | MMAR_4782 | narL | 6e-07 | 47.27% (55) | nitrate/nitrite response regulator protein NarL |
| M. avium 104 | MAV_5280 | - | 3e-07 | 45.45% (55) | two-component system response regulator |
| M. thermoresistible (build 8) | TH_1532 | - | 9e-10 | 42.86% (56) | two-component system response regulator |
| M. ulcerans Agy99 | MUL_0351 | narL | 2e-07 | 47.27% (55) | nitrate/nitrite response regulator protein NarL |
| M. vanbaalenii PYR-1 | Mvan_5906 | - | 4e-10 | 50.00% (56) | two component LuxR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4782|M.marinum_M -----------------MDVTKTEKVRVVVGDDHPLFREGVVRALSLSGS
MUL_0351|M.ulcerans_Agy99 -----------------MDVTKTEKVRVVVGDDHPLFREGVVRALSLSGS
MAV_5280|M.avium_104 ------------------MTDPAPEITVLLVDDQDLVRSGLRRILRRKDG
Mflv_0968|M.gilvum_PYR-GCK -----------------MTG-----LRVVLAEDAPLLRAGIVTVLE-SDG
Mvan_5906|M.vanbaalenii_PYR-1 --------------MPDLTSRGGERLRIVLAEDAPLLRAGIVTVLE-SDG
TH_1532|M.thermoresistible__bu -------------------------MRIVIAEDSALLRAGIERILT-DAG
MAB_4498c|M.abscessus_ATCC_199 ----------------------MSAIRVLVADDDTLLREGVASVLE-KAG
Mb1652|M.bovis_AF2122/97 ------------MTGPTTDADAAVPRRVLIAEDEALIRMDLAEMLR-EEG
Rv1626|M.tuberculosis_H37Rv ------------MTGPTTDADAAVPRRVLIAEDEALIRMDLAEMLR-EEG
MLBr_01286|M.leprae_Br4923 ------------MTGPTTDADAATPHRVLIAEDEALIRLDLAEMLR-DEG
MSMEG_2806|M.smegmatis_MC2_155 MCGYRDAVRPYARLRLCRRREELSVATVVVADDHSLIREGVVRVLE-RGG
::: :* *.* .: * .
MMAR_4782|M.marinum_M VNVVGEADDGAAALELIKTHLPDVALLDYRMP---RMDGAQVAAAVRSNE
MUL_0351|M.ulcerans_Agy99 VNVVGEADDGAAALELIETHLPDVALLDYRMP---RMDGAQVAAAVRSNG
MAV_5280|M.avium_104 FVIVAECADGDEVPAAVAAHRPDVVVMDLRMR---RVDG--IEATRRLGG
Mflv_0968|M.gilvum_PYR-GCK HQVCAAVESADELRAAVRAHNPDLVVTDVRMPPTHTDEGIRVAIELRRAT
Mvan_5906|M.vanbaalenii_PYR-1 HHVCAAVESADELRDAARAHDPDVVVTDVRMPPTHTDEGIRVAVDLRRTT
TH_1532|M.thermoresistible__bu HEVVAGVPDATDLLRLVNELRPDLAIVDVRMPPTFTDEGIRAAALLRSQN
MAB_4498c|M.abscessus_ATCC_199 FEVVGRAGDATALLELVRTESPNLVVVDIRMPPTHTTEGLEAAQKIRGEH
Mb1652|M.bovis_AF2122/97 YEIVGEAGDGQEAVELAELHKPDLVIMDVKMP---RRDGIDAASEIAS-K
Rv1626|M.tuberculosis_H37Rv YEIVGEAGDGQEAVELAELHKPDLVIMDVKMP---RRDGIDAASEIAS-K
MLBr_01286|M.leprae_Br4923 YDIVGEAADGQEAVELAERHKPDLVIMDVKMP---RRDGIDAASEIAS-K
MSMEG_2806|M.smegmatis_MC2_155 HRVLAVAADAVELLVQVDIHRPDVVVTDIEMP------------------
: . .. *::.: * .*
MMAR_4782|M.marinum_M LPTRVLLISAHDESAIVYQALQ------QGAAGFLLKD--STRTEIVKAV
MUL_0351|M.ulcerans_Agy99 LPTRVLLISAHDESAIVYQALQ------QGAAGFLLKD--STRTEIVKAV
MAV_5280|M.avium_104 RPP-VLALTTFNEDELLSGALR------AGAAGFVLKD--SSAEELIRAV
Mflv_0968|M.gilvum_PYR-GCK PGLPVVILSQYTSVGSLDQLLDRTEDAGRAGLGYLLKDRVAHVRHFLDAV
Mvan_5906|M.vanbaalenii_PYR-1 PGLPVVILSQYTTVGSLDQLLDRTAATGRGGLGYLLKDRVAHVRHFLDAV
TH_1532|M.thermoresistible__bu PESPVLVLSHYVEERYAADLIA----SDTKGFGYLLKDRVADVPAFLDAV
MAB_4498c|M.abscessus_ATCC_199 PGIGVLVLSAHVEVEYAAELIT-----SGDGVGYLLKSRIGDVGEFADAC
Mb1652|M.bovis_AF2122/97 RIAPIVVLTAFSQRDLVERARD------AGAMAYLVKP--FSISDLIPAI
Rv1626|M.tuberculosis_H37Rv RIAPIVVLTAFSQRDLVERARD------AGAMAYLVKP--FSISDLIPAI
MLBr_01286|M.leprae_Br4923 RIAPIVVLTAFSQRDLVERARD------AGAMAYLVKP--FTISDLIPAI
MSMEG_2806|M.smegmatis_MC2_155 -----------------------------------------PSVARLKFV
MMAR_4782|M.marinum_M LDCAKGRDVVAPSLVGGLAGEIRQRAAP----MAPVLSAREREVLHRIAK
MUL_0351|M.ulcerans_Agy99 LDCAKGRDVVAPSLVGGLAGEIRQRAAP----MAPVLSAREREVLHRIAK
MAV_5280|M.avium_104 RAVARGEGYLDPAVTSRVLTTYRKAAPGPHGAAIAELTTRERDVLTLIGK
Mflv_0968|M.gilvum_PYR-GCK RDIASGATIIDPDIVVALVGASRRRGVL------ATLTPREQEVLGLMAQ
Mvan_5906|M.vanbaalenii_PYR-1 RDIAAGATIIDPEIVVALVGASRRRGVL------AALTPREEEVLGLMAQ
TH_1532|M.thermoresistible__bu EVVGGGGTVLDPEVVSQILVRSHRRSAL------DELTAREHDVLRLMAE
MAB_4498c|M.abscessus_ATCC_199 TRVAGGGSIVDPELVRELLSARRKDDPL------SALSQRELEVLELMAQ
Mb1652|M.bovis_AF2122/97 ELAVSRFREIT-ALEGEVATLSERLETR-------KLVERAKGLLQTKH-
Rv1626|M.tuberculosis_H37Rv ELAVSRFREIT-ALEGEVATLSERLETR-------KLVERAKGLLQTKH-
MLBr_01286|M.leprae_Br4923 ALAMSRFSELT-ALEREVATLADRLETR-------KLVERAKGLLQVKQ-
MSMEG_2806|M.smegmatis_MC2_155 YECGAG-------FRRRSWS------------------------------
.
MMAR_4782|M.marinum_M GQSIPAIAGELYVAPSTVKTHVQRLYEKLGVSDRAAAVAEAMRQGLLD--
MUL_0351|M.ulcerans_Agy99 GQSIPAIAGELYVAPSTVKTHVQRLYEKLGVSDRAAAVAEAMRQGLLD--
MAV_5280|M.avium_104 GLSNSEIADELCISGVTVKSHIGRIFGKLDLRDRAAAIVYAYDNGIVAPR
Mflv_0968|M.gilvum_PYR-GCK GLTNPQIAQRLVVSEAAVRKHVGNIFAKLPLSADG--DRRVLAVLTYLNE
Mvan_5906|M.vanbaalenii_PYR-1 GLTNPQIAQRLVVSEAAVRKHVGGIFAKLPLSDDG--DRRVLAVLTYLNE
TH_1532|M.thermoresistible__bu GKTNSAIAGALHISIGSAEKHIASIFTKLGLEPDESENRRVLAVLRYLES
MAB_4498c|M.abscessus_ATCC_199 GLSNSGIAGRLWITESTVEKHVHSVMRRLDLGHADEYHRRVLAVLAFLTH
Mb1652|M.bovis_AF2122/97 GMTEPDAFKWIQRAAMDRRTTMKRVAEVVLETLGTPKDT-----------
Rv1626|M.tuberculosis_H37Rv GMTEPDAFKWIQRAAMDRRTTMKRVAEVVLETLGTPKDT-----------
MLBr_01286|M.leprae_Br4923 GMTEPEAFKWIQRAAMDRRTTMKRVAEVVLETLDTPKDT-----------
MSMEG_2806|M.smegmatis_MC2_155 ----------------------TRMGGRVGSGPR----------------
: :
MMAR_4782|M.marinum_M -
MUL_0351|M.ulcerans_Agy99 -
MAV_5280|M.avium_104 -
Mflv_0968|M.gilvum_PYR-GCK H
Mvan_5906|M.vanbaalenii_PYR-1 S
TH_1532|M.thermoresistible__bu -
MAB_4498c|M.abscessus_ATCC_199 R
Mb1652|M.bovis_AF2122/97 -
Rv1626|M.tuberculosis_H37Rv -
MLBr_01286|M.leprae_Br4923 -
MSMEG_2806|M.smegmatis_MC2_155 -