For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TYPPSNPPASPPPGPDGPWQQPPQAYPQYGDAQQGYQPGSAPGAQPAYGAPQYDPQAYGYPQQPAYGQQQ YGQPQYGQPQYGQPQYGQPQYGQQPYGYGPQGYPQPQPPSSNKKGLIIGGAIAAVVVLVAIIALVVLLLV PSQSSDQKAIQQLLKDVGSTSNFSTALDNYFCAGDQALFDMSALEELGIDPSLIDSPPMETPDANATIGD ITVDGDKATAKIESRGSTGTMHFRKESGEWKVCMSDSPTLSNFPGFN*
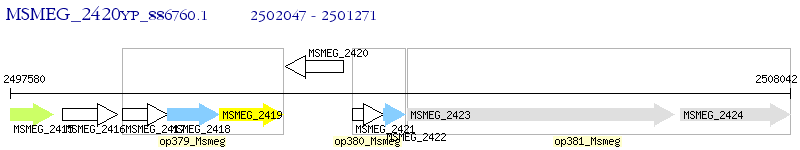
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2420 | - | - | 100% (258) | hypothetical protein MSMEG_2420 |
| M. smegmatis MC2 155 | MSMEG_3884 | - | 3e-21 | 44.32% (176) | hypothetical protein MSMEG_3884 |
| M. smegmatis MC2 155 | MSMEG_0035 | - | 3e-15 | 44.23% (104) | FHA domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2118c | - | 3e-12 | 35.40% (161) | hypothetical protein Mb2118c |
| M. gilvum PYR-GCK | Mflv_3096 | - | 1e-16 | 37.72% (167) | hypothetical protein Mflv_3096 |
| M. tuberculosis H37Rv | Rv2091c | - | 3e-12 | 35.40% (161) | hypothetical protein Rv2091c |
| M. leprae Br4923 | MLBr_01334 | - | 1e-08 | 33.13% (166) | hypothetical protein MLBr_01334 |
| M. abscessus ATCC 19977 | MAB_2190 | - | 7e-15 | 42.57% (148) | hypothetical protein MAB_2190 |
| M. marinum M | MMAR_3077 | - | 1e-15 | 37.10% (186) | hypothetical protein MMAR_3077 |
| M. avium 104 | MAV_2412 | - | 1e-13 | 42.17% (166) | hypothetical protein MAV_2412 |
| M. thermoresistible (build 8) | TH_3511 | - | 4e-19 | 36.62% (142) | PUTATIVE - |
| M. ulcerans Agy99 | MUL_2323 | - | 7e-16 | 37.10% (186) | hypothetical protein MUL_2323 |
| M. vanbaalenii PYR-1 | Mvan_3439 | - | 4e-18 | 40.41% (146) | hypothetical protein Mvan_3439 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2118c|M.bovis_AF2122/97 MSGP----QGSDPRQPW-------QPPGQGAD----HSSDPTVAAGYPWQ
Rv2091c|M.tuberculosis_H37Rv MSGP----QGSDPRQPW-------QPPGQGAD----HSSDPTVAAGYPWQ
MMAR_3077|M.marinum_M MSGP----QGPDPRQSW-------QPPGQGGD----HSSDPTMAAGSPWQ
MUL_2323|M.ulcerans_Agy99 MSGP----QGPDPRQSW-------QPPGQGGD----HSSDPTMAAGSPWQ
MAV_2412|M.avium_104 ------------------------------------------MAP--TWQ
MLBr_01334|M.leprae_Br4923 MSEP----QGSDPGKQW-------QSPGEGVEN---HSSDQPTQAASPWQ
Mflv_3096|M.gilvum_PYR-GCK MSGP----QGSDPTQPWSG-----QQPEQGTDQ---PAGD--AAANQGWQ
Mvan_3439|M.vanbaalenii_PYR-1 MSGP----QGSDPTQPWPG-----QQPEQGTDQ---PAGD--SAANQGWQ
MAB_2190|M.abscessus_ATCC_1997 MASLPLMAHKVEQTLRWSVSTLLHTGKKQGRDTGGTMSGPQGTEPGGQWA
MSMEG_2420|M.smegmatis_MC2_155 MTYP--------------------PSNPPASPPPGPDGPWQQPPQAYPQY
TH_3511|M.thermoresistible__bu VVIV--------------------LVLVIAVTS-----------------
Mb2118c|M.bovis_AF2122/97 QQ------PTQEATWQAPAYTP-----QYQQPADP-AY--PQQYPQPTPG
Rv2091c|M.tuberculosis_H37Rv QQ------PTQEATWQAPAYTP-----QYQQPADP-AY--PQQYPQPTPG
MMAR_3077|M.marinum_M Q-------PGQEGSWQPPSYPAA-DYQQYQQPVEP-AY--PQQYP-AAPG
MUL_2323|M.ulcerans_Agy99 Q-------PGQEGSWQPPSYPAA-DYQQYQQPVEP-AY--PQQYP-AAPG
MAV_2412|M.avium_104 DH------ATQEASWQAPAYTPPAEYPQYQQPGEQ-AY--PQPYP-QQQG
MLBr_01334|M.leprae_Br4923 QQP-----STQDSTWHPPAYASP-ECYNYPQLTEP-VY--PHQYPSATPG
Mflv_3096|M.gilvum_PYR-GCK PPS------TEQPAWQPPAYQPQ-QYPQYQQPTQPPTYS-PQQYPPGDQY
Mvan_3439|M.vanbaalenii_PYR-1 PPTPGAGQQPEQPSWQPPAYTPQ-QYPQYQQPAQPPTYS-PQQYP-ADQY
MAB_2190|M.abscessus_ATCC_1997 PQP-----GQGQDPWQTPQSSDP--AQQGEQGWQQPAYGQGQQYP-QYQQ
MSMEG_2420|M.smegmatis_MC2_155 GDAQQGYQPGSAPGAQPAYGAPQYDPQAYGYPQQPAYGQQQYGQPQYGQP
TH_3511|M.thermoresistible__bu -------KSGSGPANRAAG--------ALGNSEE----------------
:.. :
Mb2118c|M.bovis_AF2122/97 YA-QPEQFGA----QPTQLGVP------GQYG---QYQQPGQYGQPGQYG
Rv2091c|M.tuberculosis_H37Rv YA-QPEQFGA----QPTQLGVP------GQYG---QYQQPGQYGQPGQYG
MMAR_3077|M.marinum_M YG-QSD-FGS----QPTQFGAPPQFGQPGQYGQPGQYGQPGQYGQPGQPG
MUL_2323|M.ulcerans_Agy99 YG-QSD-FGS----QPTRFGAPPQFGQPGQYGQPGQYGQPGQYGQPGQPG
MAV_2412|M.avium_104 YA-QPGQYGQY--GQPGQYGQP------GQYAQPGQYPQPGQYPQPGQYP
MLBr_01334|M.leprae_Br4923 YG-QPGYFGA----QFSQCGIP------------GQYPQSG---SPGQYG
Mflv_3096|M.gilvum_PYR-GCK GQPQTGEYNPAAYPPPGQYGQQ------PGYG---QYGQQPQYGQPGQYG
Mvan_3439|M.vanbaalenii_PYR-1 GQ-QPAEYSPAGYPPPGQYGQP------GGYG---QQYGQP-YGQPGQYG
MAB_2190|M.abscessus_ATCC_1997 PGFPGGGFGD-----PSQFGQQPG----QGFGEPSQFGQPGAYGQQGQYG
MSMEG_2420|M.smegmatis_MC2_155 QYGQPQYGQPQYGQQPYGYGPQ-------------GYPQPQPPSSNKKGL
TH_3511|M.thermoresistible__bu --------------------------------------------------
Mb2118c|M.bovis_AF2122/97 -QPG---QYAP-PGQYPGQYGPYGQSGQGSKRSVAVIGGVIAVMAVLFIG
Rv2091c|M.tuberculosis_H37Rv -QPG---QYAP-PGQYPGQYGPYGQSGQGSKRSVAVIGGVIAVMAVLFIG
MMAR_3077|M.marinum_M -QPG---QYGQ-PGQYPGQFSPYDQSGQGSKRSMALIGGVVGGIALLFVV
MUL_2323|M.ulcerans_Agy99 -QPG---QYGQ-PGQYPGQFSPYDQSGQGSKRSMALIGGVVGGIALLFVV
MAV_2412|M.avium_104 -QPGQYGQYGQ-PGQYPQQYQPYEQPGK--KRPVGLIAGVAGGIALLLIA
MLBr_01334|M.leprae_Br4923 -PPG---QYGP-PGQYSQQFQPYEQPGT--KGFVALIGSIAGVIGVLIFA
Mflv_3096|M.gilvum_PYR-GCK QQPG---QYGQYPGAP---GYPGQGGDEGSKRSMAVIGGVIGALVAVILA
Mvan_3439|M.vanbaalenii_PYR-1 QQPGQAGQYGQYPGAPGAPGYPAPGSEEGSKRSLAVIGGVVGVLAAVVIA
MAB_2190|M.abscessus_ATCC_1997 QQGAYPGQYGQPQGQPFPQFGAGK--PQRSTKTIAIIGGIVA--AAVVLI
MSMEG_2420|M.smegmatis_MC2_155 IIGGAIAAVVVLVAIIALVVLLLVPSQSSDQKAIQQL--LKDVGSTSNFS
TH_3511|M.thermoresistible__bu -------------AAIRAMIDSLAASESTDT------------------S
.
Mb2118c|M.bovis_AF2122/97 AVLILGFWAPGFFVTTKLDVIKAQAGVQQVLTDETTGYG-AKNVKDVKCN
Rv2091c|M.tuberculosis_H37Rv AVLILGFWAPGFFVTTKLDVIKAQAGVQQVLTDETTGYG-AKNVKDVKCN
MMAR_3077|M.marinum_M VILVLGFWVPGFFVTTKLDVSKANAGVQQVLTDETNGYG-AKNVKDVKCN
MUL_2323|M.ulcerans_Agy99 VILVLGFWVPGFFVTTKLDVSKANAGVQQVLTDETNGYG-AKNVKDVKCN
MAV_2412|M.avium_104 VVLVLGFWQPGFFVTTKLDVNKAQAGVQQVLSDETNGYG-AKNVKDVKCN
MLBr_01334|M.leprae_Br4923 AILVTGFLWPAWLVTTKLDVNKAQASVQQVLTDETNGYG-AKNVKDVKCN
Mflv_3096|M.gilvum_PYR-GCK VVLVTGFWKPGFFVTTKLDVDAAQTGVRQILTDETNGYG-AKNVNDVTCN
Mvan_3439|M.vanbaalenii_PYR-1 AVLVLGFWKPGFFVTTKLDVNAAQNGVKQILTDETNGYG-ATNVNDVKCN
MAB_2190|M.abscessus_ATCC_1997 VLLVTAFLAPGWAVTTTLDVNKAQDGVTKILTDETNGYG-AKNVKDVKCN
MSMEG_2420|M.smegmatis_MC2_155 TALDNYFCAGDQALFDMSALEELGIDPSLIDSPPMETPDANATIGDITVD
TH_3511|M.thermoresistible__bu GLLREYFCAGDRELFDSFDLG--GLDLPSTAGPATDVP----EISDIRVN
* * : : . : *: :
Mb2118c|M.bovis_AF2122/97 NGSDPTVKKGATFECTVSIDGTSKRVTVTFQDNKGTYEVGRPQ-
Rv2091c|M.tuberculosis_H37Rv NGSDPTVKKGATFECTVSIDGTSKRVTVTFQDNKGTYEVGRPQ-
MMAR_3077|M.marinum_M DGTDPTVKKGATFDCTVSIDGASKHVTVTFQDNKGTYEVGRPQ-
MUL_2323|M.ulcerans_Agy99 DGTDPTVKKGATFDCTVSIDGASKHVTVTFQDNKGTYEVGRPQ-
MAV_2412|M.avium_104 HGQNPTVKKGATFDCDVSIDGAPKHVTVTFQDNKGTYEVGRPQ-
MLBr_01334|M.leprae_Br4923 NGADPTVKKGDTFDCSVSIDGMQKRVTVTFQDDKGTYEVGRPQ-
Mflv_3096|M.gilvum_PYR-GCK GGVSPTVKKGDTFTCEVSIDGTKRQVTVTFQDDDGTYEVGRPR-
Mvan_3439|M.vanbaalenii_PYR-1 NGVSPTVKKGDTFNCEVSIDGTKRQVTVTFQDDNGTYEVGRPK-
MAB_2190|M.abscessus_ATCC_1997 DGNNPEVKKGATFTCDVSIDGTKRQVTVTFQNDKGTYEVGRPK-
MSMEG_2420|M.smegmatis_MC2_155 GDKATAKIESRGSTGTMHFRKESGEWKVCMSDSPTLSNFPGFN-
TH_3511|M.thermoresistible__bu GDKATARASVKGVASTMHFRKEGGKWKFCLTDAPEFSEMPRFGR
. . . : : . .. : : :.