For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
DLKWSKTDQEFRDEVRGFLAEKLTPDLREAGRLQTSVYSEHEASMEWQRILHERGWAAPAWPVEHGGCDW SLTQHYIFSRESTLAGAPALSPMGIRMVAHAIIKFGTEEQKSYFLPRILTGEVFFCQGYSEPEAGSDLAA LSMSAVSDGDDLVCTGSKIWTTHAQEANWMFALVRTTRGAKKQQGITFVLIDMTSPGIQIRPLVMTSGEE VQNQVFFDEVRVPKKNVIGAIDDGWTVAKYLLEFERGGGASSPALQVLGEEIATVAARQPGPSGGRLIDD PAFSRKLADARIRTDVLEILEYQVLAAVAEGRNPGTASSMLKVLSTELSQILTELSMEAAGPRGRAYQPH ATRPGGPVAEFEPPADGYVSGEPWQAVAPLRYFNDRAGSIYAGSNEIQRNILAKAALGL*
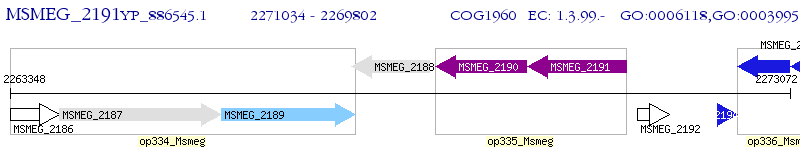
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2191 | - | - | 100% (410) | acyl-CoA dehydrogenase family protein |
| M. smegmatis MC2 155 | MSMEG_6511 | - | 1e-71 | 39.09% (417) | acyl-CoA dehydrogenase domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_6012 | - | 3e-51 | 34.13% (416) | putative acyl-CoA dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1969c | fadE17 | 5e-65 | 36.28% (419) | acyl-CoA dehydrogenase FADE17 |
| M. gilvum PYR-GCK | Mflv_4379 | - | 0.0 | 85.85% (410) | acyl-CoA dehydrogenase domain-containing protein |
| M. tuberculosis H37Rv | Rv1934c | fadE17 | 5e-65 | 36.28% (419) | acyl-CoA dehydrogenase FADE17 |
| M. leprae Br4923 | MLBr_00737 | fadE25 | 5e-19 | 22.87% (411) | putative acyl-CoA dehydrogenase |
| M. abscessus ATCC 19977 | MAB_2495 | - | 1e-178 | 74.63% (410) | acyl-CoA dehydrogenase |
| M. marinum M | MMAR_3737 | fadE17_1 | 5e-77 | 42.09% (411) | acyl-CoA dehydrogenase FadE17_1 |
| M. avium 104 | MAV_1766 | - | 5e-74 | 39.66% (411) | acyl-CoA dehydrogenase domain-containing protein |
| M. thermoresistible (build 8) | TH_4368 | - | 0.0 | 84.88% (410) | PUTATIVE acyl-CoA dehydrogenase family protein |
| M. ulcerans Agy99 | MUL_3681 | fadE17_1 | 3e-77 | 42.09% (411) | acyl-CoA dehydrogenase FadE17_1 |
| M. vanbaalenii PYR-1 | Mvan_1965 | - | 0.0 | 86.34% (410) | acyl-CoA dehydrogenase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1969c|M.bovis_AF2122/97 -MDVSYPPEAEAFR--DRIREFVAEHLPPGWPGPGALPPHERE--EFARH
Rv1934c|M.tuberculosis_H37Rv -MDVSYPPEAEAFR--DRIREFVAEHLPPGWPGPGALPPHERE--EFARH
Mflv_4379|M.gilvum_PYR-GCK -MDLGWSATDLEFR--DEVRAFLEEKLAPELRQAGRLMTSVYADHDASMA
Mvan_1965|M.vanbaalenii_PYR-1 -MDLEWSATDLAFR--DEVRAFLDDRLTPELRRAGRLMTSVYADHDASMA
MSMEG_2191|M.smegmatis_MC2_155 -MDLKWSKTDQEFR--DEVRGFLAEKLTPDLREAGRLQTSVYSEHEASME
TH_4368|M.thermoresistible__bu -MDLQWSPADLAFR--DEVRAFLDEKLTPGLRRAGRLQTSVYSDHEASME
MAB_2495|M.abscessus_ATCC_1997 -MDLAFSAEEEQFR--TEVREFLDANLTAELRAAGRLQTSVYSDHEASMR
MMAR_3737|M.marinum_M -MKLALTPDEAAFR--DELRTFYTTRIPEDLRELVR-QGQSPS-RDEIVT
MUL_3681|M.ulcerans_Agy99 -MKLALTPDEAAFR--DELRTFYTTRIPEDLRELVR-QGQSPS-RDEIVT
MAV_1766|M.avium_104 -MQLALTPEEAAFR--DELRTIYTTKIPEELRERVR-RGSAEVNRDDIVT
MLBr_00737|M.leprae_Br4923 MVGWSGNPLFDLFKLPEEHNELRATIRALAEKEIAPHAADVDQRARFPEE
: *: . . : .
Mb1969c|M.bovis_AF2122/97 WRRALAGAGLVAVSWPTEYGGGGLSPMEQVVLAEEFARAGAPERAENDLL
Rv1934c|M.tuberculosis_H37Rv WRRALAGAGLVAVSWPTEYGGGGLSPMEQVVLAEEFARAGAPERAENDLL
Mflv_4379|M.gilvum_PYR-GCK WQAILHERGWAAPAWPVAHGGCDWSLTQHYIFSRESTLAGAPSLSP---M
Mvan_1965|M.vanbaalenii_PYR-1 WQQILHERGWAAPAWPVAHGGCDWSLTQHYIFSRESTLAGAPSLSP---M
MSMEG_2191|M.smegmatis_MC2_155 WQRILHERGWAAPAWPVEHGGCDWSLTQHYIFSRESTLAGAPALSP---M
TH_4368|M.thermoresistible__bu WQRILHERGWAAPAWPVEYGGCDWTLTQHYIFERETTLAGAPSLSP---M
MAB_2495|M.abscessus_ATCC_1997 WQAILHRKGWAAPAWPVKYGGQPWSLAQHYIFSVESMVAGAPSLSP---M
MMAR_3737|M.marinum_M CHKILNDHGLAVPNWPVEWGGKDWTPTQHQIWADEMQLASVPEPLN---F
MUL_3681|M.ulcerans_Agy99 CHKILNDHGLAVPNWPVEWGGKDWTPTQHQIWADEMQLASVPEPLN---F
MAV_1766|M.avium_104 SHKILHEHGLAVPNWPVEWGGKDWTPTQHQIWADEMQLACVPEPLN---F
MLBr_00737|M.leprae_Br4923 ALAALNASGFNAIHVPEEYGGQGADSVAACIVIEEVARVDASASLIP--A
* * . * ** : * . ..
Mb1969c|M.bovis_AF2122/97 GIDLLGNTLIALGSEAQKRHFLPRILSGEHRWCQGFSEPEAGSDLASVRT
Rv1934c|M.tuberculosis_H37Rv GIDLLGNTLIALGSEAQKRHFLPRILSGEHRWCQGFSEPEAGSDLASVRT
Mflv_4379|M.gilvum_PYR-GCK GIRMVAHAIIKFGTDEQKDFFLPRILTGEVFFCQGYSEPEAGSDLAALSM
Mvan_1965|M.vanbaalenii_PYR-1 GIRMVAHAIIRFGSDEQKNYFLPRILTGEVFFCQGYSEPEAGSDLAALSM
MSMEG_2191|M.smegmatis_MC2_155 GIRMVAHAIIKFGTEEQKSYFLPRILTGEVFFCQGYSEPEAGSDLAALSM
TH_4368|M.thermoresistible__bu GIKMVAHAIIAYGTQEQKDYFLPRILTGEVFFCQGYSEPEAGSDLAALSM
MAB_2495|M.abscessus_ATCC_1997 GIRMVAHAIIKFGSPEQKEFFLPRILTGEVFFCQGYSEPESGSDLASLQM
MMAR_3737|M.marinum_M NTKMVGPVIAEFGSQEIKQRFLPPTASLDIWWCQGFSEPEAGSDLASLRT
MUL_3681|M.ulcerans_Agy99 NTKMVGPVIAEFGSQEIKQRFLPPTASLDIWWCQGFSEPEAGSDLASLRT
MAV_1766|M.avium_104 NTKMVGPVIAEFGSEEVKQRFLPPTASLDIWWCQGFSEPDAGSDLASLRT
MLBr_00737|M.leprae_Br4923 VNKLGTMGLILRGSEELKKQVLPSLAAEGAMASYALSEREAGSDAASMRT
: : *: * .** : . . ** ::*** *::
Mb1969c|M.bovis_AF2122/97 RGVLDGDEWVINGHKIWTSAGTTANWIFLLARTD-PSAAKHRGLSFLLVP
Rv1934c|M.tuberculosis_H37Rv RGVLDGDEWVINGHKIWTSAGTTANWIFLLARTD-PSAAKHRGLSFLLVP
Mflv_4379|M.gilvum_PYR-GCK AARDDGDHLVCTGSKIWTTHAQEANWMFALVRTT-RTDKKQRGITFVLID
Mvan_1965|M.vanbaalenii_PYR-1 AARDDGDHLVCTGSKIWTTHAREANWMFALVRTT-RSEKKQRGITFVLID
MSMEG_2191|M.smegmatis_MC2_155 SAVSDGDDLVCTGSKIWTTHAQEANWMFALVRTT-RGAKKQQGITFVLID
TH_4368|M.thermoresistible__bu AAVDDGDDLICSGSKIWTTHAVEANWMFALVRTS-RSDKKQKGITFLLID
MAB_2495|M.abscessus_ATCC_1997 SAVADGDDFVLNGSKIWTTHAREANWMFALVRTT-KGPKKQLGITFILID
MMAR_3737|M.marinum_M TAVRDGDSYVVNGQKTWTTLGQYADWIFCLVRTDPQAPKRQAGISFLLID
MUL_3681|M.ulcerans_Agy99 TAVRDGDSYVVNGQKTWTTLGQYADWIFCLVRTDPQAPKRQAGISFLLID
MAV_1766|M.avium_104 TAVRDGDHYIVNGQKTWTTLGQYADWIFCLVRTDPQAPKRQAGISFLLFD
MLBr_00737|M.leprae_Br4923 RAKADGDDWILNGFKCWITNGGKSTWYTVMAVTD--PDKGANGISAFIVH
. *** : .* * * : . : * :. * *:: .:.
Mb1969c|M.bovis_AF2122/97 MDQPGVVVRPIVNAAGH--SSFSEVFLTDARTSAGNVVGRVGDGWSTAMT
Rv1934c|M.tuberculosis_H37Rv MDQPGVVVRPIVNAAGH--SSFSEVFLTDARTSAGNVVGRVGDGWSTAMT
Mflv_4379|M.gilvum_PYR-GCK MATPGIEIRPLVMTSGE--EVQNQVFFDEVRVPKSNVIGQIDDGWTVAKY
Mvan_1965|M.vanbaalenii_PYR-1 MSTPGIEIRPLVMTSGE--EVQNQVFFDDVRVPKANVIGEIDDGWTVAKY
MSMEG_2191|M.smegmatis_MC2_155 MTSPGIQIRPLVMTSGE--EVQNQVFFDEVRVPKKNVIGAIDDGWTVAKY
TH_4368|M.thermoresistible__bu MTSPGIEIRPLVMTSGE--VVQNQVFFDEVRVPKRNVVGKIDEGWTVAKY
MAB_2495|M.abscessus_ATCC_1997 FTSPGIDVKPLVMSSGE--EVQNVVFFDNVRVPRANVLGEVDQGWTVAKY
MMAR_3737|M.marinum_M MATPGITLRPIKTIDGG--HEVNEVFFSDVRVPADQLVGQENQGWTYAKF
MUL_3681|M.ulcerans_Agy99 MATPGITLRPIKTIDGG--HEVNEVFFSDVRVPADQLVGQENQGWTYAKF
MAV_1766|M.avium_104 MKSPGITLRPIKTIDGG--HEINEVFFEDVRVPADQLVGEENQGWTYAKF
MLBr_00737|M.leprae_Br4923 KDDEGFSIGPKEKKLGIKGSPTTELYFDKCRIPGDRIIGEPGTGFKTALA
*. : * * . ::: . * . .::* . *:. *
Mb1969c|M.bovis_AF2122/97 LLGFERGSH-IATAAIDFERDLQRLCELARDR----GLHTDPRVRDGLAW
Rv1934c|M.tuberculosis_H37Rv LLGFERGSH-IATAAIDFERDLQRLCELARDR----GLHTDPRVRDGLAW
Mflv_4379|M.gilvum_PYR-GCK LLEFERGGG-AVAPGLQVMAENIATVAADQPGPGGGRLLDDPAFARKLAD
Mvan_1965|M.vanbaalenii_PYR-1 LLEFERGGG-ASAPALQVMVEEITTAAAGQPGPSGGRLLDDPAFARKLAD
MSMEG_2191|M.smegmatis_MC2_155 LLEFERGGG-ASSPALQVLGEEIATVAARQPGPSGGRLIDDPAFSRKLAD
TH_4368|M.thermoresistible__bu LLEFERGGS-ATSPGLQVMADQIATAAAEQPGPGGTRLIDDPGFSRRLAD
MAB_2495|M.abscessus_ATCC_1997 LLEFERGGG-AVAPMLQVGLDQLSVDARSVPAENG-TLADDPVFCAKLAD
MMAR_3737|M.marinum_M LLGNERTGI-AGVGRTKVRLAEVKKHAANNG------LLDDPLFAARLAE
MUL_3681|M.ulcerans_Agy99 LLGNERTGI-AGVGRTKVRLAEVKKHAANNG------LLDDPLFAARLAE
MAV_1766|M.avium_104 LLGNERTGI-AGVGRTKVRLAEVKKHAAATG------VLDDPLFAARLAE
MLBr_00737|M.leprae_Br4923 TLDHTRPTIGAQAVGIAQGALDAAIVYTKDRKQFGESISTFQSIQFMLAD
* * : . **
Mb1969c|M.bovis_AF2122/97 CYARVQIMRYRGYRDLTLALTGR-PPGAEAAITKVIWSEYFRRYTDLAVE
Rv1934c|M.tuberculosis_H37Rv CYARVQIMRYRGYRDLTLALTGR-PPGAEAAITKVIWSEYFRRYTDLAVE
Mflv_4379|M.gilvum_PYR-GCK VRIRTEVLEILEYQVLAVVAGGG-NPGAKSSMLKVLGTELSQAITELAME
Mvan_1965|M.vanbaalenii_PYR-1 VRIRTEVLEILEYQVLAAVAEGG-NPGAKSSMLKVLSTELSQAITELAME
MSMEG_2191|M.smegmatis_MC2_155 ARIRTDVLEILEYQVLAAVAEGR-NPGTASSMLKVLSTELSQILTELSME
TH_4368|M.thermoresistible__bu ARIRTEVLEILEYQVLATVAAGK-NPGSASSMLKVLSTELSQTLTELAME
MAB_2495|M.abscessus_ATCC_1997 LAIRVDVLEVLEHRVLAVVAGGG-NPGAASSMLKILGTELSQALTTLTLE
MMAR_3737|M.marinum_M AENELLALELTQARVVTDSADG--KPNPASSVLKLRGSQLQQVATELLVE
MUL_3681|M.ulcerans_Agy99 AENELLALELTQARVVTDSADG--KPNPASSVLKLRGSQLQQVATELLVE
MAV_1766|M.avium_104 AENELLALELTQSRVTTDSANG--QPNPASSVLKLRGSQLQQVATELLVE
MLBr_00737|M.leprae_Br4923 MAMKVEAARLIVYAAAARAERGEPDLGFISAASKCFASDIAMEVTTDAVQ
. . : * . :: * :: * ::
Mb1969c|M.bovis_AF2122/97 ILG--LEALGPRGPGNGGARLVP------EAGTPNSPACWMDELLYARAA
Rv1934c|M.tuberculosis_H37Rv ILG--LEALGPRGPGNGGARLVP------EAGTPNSPACWMDELLYARAA
Mflv_4379|M.gilvum_PYR-GCK AAGPRGRVYQPHATYPGGPIAEFEPPEGDYVSGEPWQAVAPLRYFNDRAG
Mvan_1965|M.vanbaalenii_PYR-1 AAGPRGRVYQPHATCPGGPIAEFEPPADDYVSGEPWQAVAPLRYFNDRAG
MSMEG_2191|M.smegmatis_MC2_155 AAGPRGRAYQPHATRPGGPVAEFEPPADGYVSGEPWQAVAPLRYFNDRAG
TH_4368|M.thermoresistible__bu AAGPQGRAYQPHATCPGGPVSDFVPPADGYVTGEPWQAVAPLRYFNDRAG
MAB_2495|M.abscessus_ATCC_1997 AAGPRGRAYQPHVTKPGGPVVEYTPPADGYHSGELWQAVAPLHYFNDRAG
MMAR_3737|M.marinum_M VAG-------PDALPVGGAQAAG-------IASPQWAQSSAPRYLNYRKT
MUL_3681|M.ulcerans_Agy99 VAG-------PDALPVGGAQAAG-------IASPQWAQSSAPRYLNYRKT
MAV_1766|M.avium_104 VAG-------PDALP---AKADD-------IASPSWAQSSAPHYLNYRKT
MLBr_00737|M.leprae_Br4923 LFG---------------------------GAGYTSDFPVERFMRDAKIT
* :
Mb1969c|M.bovis_AF2122/97 TIYAGSSQIQRNVIGERLLGLPKEPRPEVLC
Rv1934c|M.tuberculosis_H37Rv TIYAGSSQIQRNVIGERLLGLPKEPRPEVLC
Mflv_4379|M.gilvum_PYR-GCK SIYAGSNEIQRNILAKAALGL----------
Mvan_1965|M.vanbaalenii_PYR-1 SIYAGSNEIQRNILAKAALGL----------
MSMEG_2191|M.smegmatis_MC2_155 SIYAGSNEIQRNILAKAALGL----------
TH_4368|M.thermoresistible__bu SIYAGSNEIQRNILAKAALGL----------
MAB_2495|M.abscessus_ATCC_1997 SIYAGSNEIQRNILAKAQLGL----------
MMAR_3737|M.marinum_M SIYGGSNEVQRNIIASTILGL----------
MUL_3681|M.ulcerans_Agy99 SIYGGSNEVQRNIIASTILGL----------
MAV_1766|M.avium_104 SIYGGSSEVQRNIIASTILGL----------
MLBr_00737|M.leprae_Br4923 QIYEGTNQIQRVVMSRALLR-----------
** *:.::** ::. *