For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTKTDNPILITGATGRHGNTGEYLVRRLREEGRAVRVLARTPSERTERLAELGAEIAIGDLHDRRTLVPA LADVDLAYFTYPIDAGVIPAAANYAAAVREVGRQPRTVVMSMGPSHPENRSDLGRAQWLAEEVMQWAGLD LLILRVAALFHENLKVLHSRSIREHGLIRNSFGPQRVAWISGRDAAELALAGLLHPQRFSETIVYIKGSE EHSHVEIAELLSELSGSPVRYETATQEQWRDDLLDLSRADGEDVVNPAMAQHISAVGHMVAHMGRTLPAD RAVLRELIGREPLPLRAFLESNLDRWSPVSV
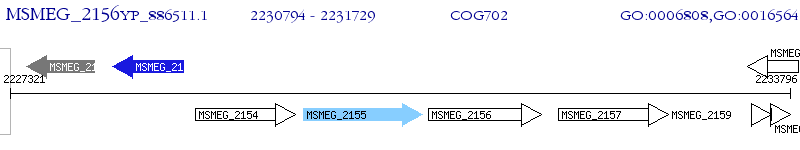
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2156 | - | - | 100% (311) | hypothetical protein MSMEG_2156 |
| M. smegmatis MC2 155 | MSMEG_3355 | - | 4e-10 | 23.78% (307) | hypothetical protein MSMEG_3355 |
| M. smegmatis MC2 155 | MSMEG_5799 | - | 1e-05 | 28.00% (150) | nucleoside-diphosphate-sugar epimerase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1635 | - | 9e-05 | 26.32% (152) | NAD-dependent epimerase/dehydratase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_4310 | - | 2e-06 | 42.25% (71) | oxidoreductase |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_0069 | - | 5e-06 | 30.97% (113) | nucleoside-diphosphate-sugar epimerase |
| M. ulcerans Agy99 | MUL_4328 | - | 7e-05 | 27.74% (155) | oxidoreductase |
| M. vanbaalenii PYR-1 | Mvan_5113 | - | 2e-07 | 30.65% (124) | NAD-dependent epimerase/dehydratase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1635|M.gilvum_PYR-GCK MPGNDVRCVVTGATG---YIGGRLVPELLARGLQVRAMARTPSKLDRTEW
Mvan_5113|M.vanbaalenii_PYR-1 MSTEQVRCLVTGATG---YIGRRLVPELLDRGHTVRAMARTPAKLDDAAW
TH_0069|M.thermoresistible__bu MP-DLPRCLVTGATG---YIGGRLIPRLLDRGHPVQAMARNPEKLAGAPW
MMAR_4310|M.marinum_M MSAGQVNCLVTGATG---YIGARLVPRLLDEGHRVRALARDPGKLADVPW
MUL_4328|M.ulcerans_Agy99 MHNRCMHVLVTGGTG---FVGGWTAKAIADAGHSVRFLVRNPAKLESSVA
MSMEG_2156|M.smegmatis_MC2_155 MTKTDNPILITGATGRHGNTGEYLVRRLREEGRAVRVLARTPSERTERLA
* ::**.** * : * *: :.* * :
Mflv_1635|M.gilvum_PYR-GCK R---DRVEVVRGDLTEADSLTAAFNGADVVYYLVHSMGT----SRDFVAE
Mvan_5113|M.vanbaalenii_PYR-1 R---DRVEVVRGDLTEPDSLASAFDGVDVIYYLVHSMGT----SRDFVAE
TH_0069|M.thermoresistible__bu R---DRVDVVQGDLTDGDSLTAAFAGVDVVYYLVHSMGS----ADDFAAE
MMAR_4310|M.marinum_M R---DRAEVVRGDLGDTDSLEAAFAGMDVVYYLVHSMGS----AKHFADE
MUL_4328|M.ulcerans_Agy99 KLGVDVSDFIIGDIKDRDLVREALTGCDAVVHSAALVATDQRQTQDMLST
MSMEG_2156|M.smegmatis_MC2_155 ELG---AEIAIGDLHDRRTLVPALADVDLAYFTYPIDAG-----------
. :. **: : : *: . * . .
Mflv_1635|M.gilvum_PYR-GCK ERRSAHNVVEAAKKAGVRRIVYLSGLHPD-RKELSRHLASRVEVG-----
Mvan_5113|M.vanbaalenii_PYR-1 EARSARNVVAAAEKAGVRRLVYLGGLHPQGKTDLSRHLASRVEVG-----
TH_0069|M.thermoresistible__bu EARSAREVVAAARRTGVRRLVYLGGLHPP-GTGLSTHLESRVEVG-----
MMAR_4310|M.marinum_M EARAAHNVVLAARRSGVRRVVYLSGLHPE-QSKLSPHLQSRMAVG-----
MUL_4328|M.ulcerans_Agy99 NMEGARNVLGQAVALGLDPIVHVSSFTALFHPGLQTLSADLPVVGGSDGY
MSMEG_2156|M.smegmatis_MC2_155 VIPAAANYAAAVREVGRQPRTVVMSMGPSHPENRSDLGRAQWLAE-----
.* : . * . : .: . . .
Mflv_1635|M.gilvum_PYR-GCK -----------EILMQSGIETVVLQAGIVIGSGSASFEMVRHLTDRLPVM
Mvan_5113|M.vanbaalenii_PYR-1 -----------DILLGSTVETVVLQAGIVVGSGSASFEMVRHLTDRLPVM
TH_0069|M.thermoresistible__bu -----------DILLDSGIETVVLQAGIVIGSGSASFEMIRHLTDRLPVM
MMAR_4310|M.marinum_M -----------DALIGSGIETIVLQAGVVVGSGSASFEMIRHLTDRLPVM
MUL_4328|M.ulcerans_Agy99 GTSKAQVEIYARGLQDAGAPVNITYPGMVLGPPVG--NQFGEAGEGVKAA
MSMEG_2156|M.smegmatis_MC2_155 -----------EVMQWAGLDLLILRVAALFH---ENLKVLHSRSIREHGL
: : : . :. : .
Mflv_1635|M.gilvum_PYR-GCK TTPKWVHNKIQPISISDVLHYLAEAATAEFGE---SRTWDIGGPDVMEYG
Mvan_5113|M.vanbaalenii_PYR-1 TTPKWVHNTIQPIAIADVLYYLAEAATAEVPE---SRTWDIGGPDVMEYG
TH_0069|M.thermoresistible__bu TTPKWVRNRIQPIAVDDVLHYLAEAATAAVPG---SRAWDVGGPDVLEYG
MMAR_4310|M.marinum_M TTPKWVHNRIQPIAIRDVLHYLVAAATATVPDTARARTWDVGGPDVLEYG
MUL_4328|M.ulcerans_Agy99 LEIRTIPGRNAAWLIIDVRDVAAVHAALLEPGRG-PRRYMVGG-HRIPVP
MSMEG_2156|M.smegmatis_MC2_155 IRNSFGPQRVAWISGRDAAELALAG--LLHPQRFSETIVYIKGSEEHSHV
*. : * .
Mflv_1635|M.gilvum_PYR-GCK DAMQGYADVAGLRRRVMVVLPFLTPTIASWWVGLVTPIPSGLARPLVESL
Mvan_5113|M.vanbaalenii_PYR-1 EAMQIYADIAHLRRRVMVVLPFLTPTIASLWVGLVTPIPSGLARPLVESL
TH_0069|M.thermoresistible__bu EAIQVYAEEAGRRRRVIVPLPWLTPRIASWWVGLVTPLPPGLAPPX---P
MMAR_4310|M.marinum_M DMMRVYAEVAGLHRRYLIVLPFLTPTIASLWVGTVTPIPPGLARPLIESL
MUL_4328|M.ulcerans_Agy99 ELAKMLGQVAETP-IVSVPIPDSVLRVAGSLLDLAGPYLPFDNPFTAAGM
MSMEG_2156|M.smegmatis_MC2_155 EIAELLSELSGSPVRYETAT---QEQWRDDLLDLSRADGEDVVNPAMAQH
: . .: : . :. .
Mflv_1635|M.gilvum_PYR-GCK ECDAICHEHDIDAVIPPPKGGLIGYREAVAKELRSEGGSS-AGRRHLLRF
Mvan_5113|M.vanbaalenii_PYR-1 ECDAVAHEHDIDTVIPPPEGGVTGYRDAVTQALRDDGTAAKGGRRHLLRF
TH_0069|M.thermoresistible__bu RCSATCRS-------PPS-------------------SSAPAWSRSGSGC
MMAR_4310|M.marinum_M ECDAVMRNSDIDTIIAPPAGGLTNYRRAVELALNRAAHGLPDATWASLQS
MUL_4328|M.ulcerans_Agy99 QYYTQMPDSDD----SPSE--------------LELGVTYRDPRTTLADT
MSMEG_2156|M.smegmatis_MC2_155 ISAVGHMVAHMGRTLPADR-------AVLRELIGREPLPLRAFLESNLDR
. ..
Mflv_1635|M.gilvum_PYR-GCK SPAASD--------------------------------------------
Mvan_5113|M.vanbaalenii_PYR-1 SKVVPQ--------------------------------------------
TH_0069|M.thermoresistible__bu SSAAS---------------------------------------------
MMAR_4310|M.marinum_M EPAEPLPSDPRWAGEIVYTDVRDATTTAAPEAVWKATERAINSRRWYSLA
MUL_4328|M.ulcerans_Agy99 VAALTA--------------------------------------------
MSMEG_2156|M.smegmatis_MC2_155 WSPVSV--------------------------------------------
.
Mflv_1635|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5113|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_0069|M.thermoresistible__bu --------------------------------------------------
MMAR_4310|M.marinum_M LAPRRRHRAPRWRVAQRTPTTLRLGAGTRVPGPTWLEVTLTPRGTAGSTY
MUL_4328|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_2156|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1635|M.gilvum_PYR-GCK ----------------------------------------
Mvan_5113|M.vanbaalenii_PYR-1 ----------------------------------------
TH_0069|M.thermoresistible__bu ----------------------------------------
MMAR_4310|M.marinum_M HQRAIFFPRGIPGRLSWLVLRHLYAAALRALARDVTAADR
MUL_4328|M.ulcerans_Agy99 ----------------------------------------
MSMEG_2156|M.smegmatis_MC2_155 ----------------------------------------