For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AINLELPRKLQAVVEKGHQGAAEMLRPISRKYDLAEHAYPVELDTLANLFEGISSAKTISFAGAEAFRDD ANGSGKDATVNGANMSALLNALEISWGDVALLLSVPRQGLGNAAISSVATPEQLERLGKDVWAAMAITEP GFGSDSAAVSTTAVLDGDEYVINGEKIFVTAGSRATHIVVWATLDKSLGRAAIKSFIVPREHPGVTVERL ERKLGIKASDTAVIRFENVRIPKDNLLGDPEIHVEKGFAGVMETFDNTRPIVAAMAVGIARAALEDLRAI LTDAGVEISYDKPAHAQSAAAAEFLRMEADWEAGYLLTVRSAWQADNNIPNSKEASMGKAKAARVASDIT LKAVEMAGTTGYSEQTLLEKWARDSKILDIFEGTQQIQQLVVARRLLGLTSAELK*
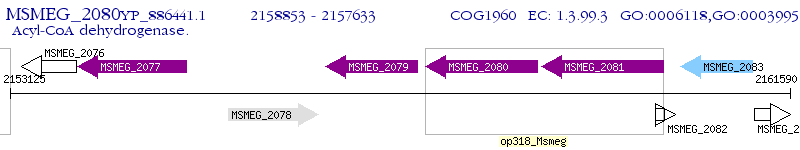
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2080 | - | - | 100% (406) | putative acyl-CoA dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_0531 | - | 1e-45 | 33.41% (422) | putative acyl-CoA dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_1821 | - | 1e-39 | 30.95% (391) | acyl-CoA dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3164 | fadE23 | 0.0 | 82.02% (406) | acyl-CoA dehydrogenase FADE23 |
| M. gilvum PYR-GCK | Mflv_4456 | - | 0.0 | 86.21% (406) | acyl-CoA dehydrogenase domain-containing protein |
| M. tuberculosis H37Rv | Rv3140 | fadE23 | 0.0 | 82.02% (406) | acyl-CoA dehydrogenase FADE23 |
| M. leprae Br4923 | MLBr_00660 | fadE23 | 0.0 | 81.28% (406) | putative acyl-CoA dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3487 | - | 0.0 | 81.77% (406) | acyl-CoA dehydrogenase |
| M. marinum M | MMAR_1509 | fadE23 | 0.0 | 79.56% (406) | acyl-CoA dehydrogenase FadE23 |
| M. avium 104 | MAV_4019 | - | 0.0 | 80.79% (406) | putative acyl-CoA dehydrogenase |
| M. thermoresistible (build 8) | TH_3136 | - | 0.0 | 89.49% (409) | PUTATIVE putative acyl-CoA dehydrogenase |
| M. ulcerans Agy99 | MUL_2432 | fadE23 | 1e-180 | 79.06% (406) | acyl-CoA dehydrogenase FadE23 |
| M. vanbaalenii PYR-1 | Mvan_1906 | - | 0.0 | 90.71% (409) | acyl-CoA dehydrogenase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_2080|M.smegmatis_MC2_155 ---MAINLELPRKLQAVVEKGHQGAAEMLRPISRKYDLAEHAYPVELDTL
Mvan_1906|M.vanbaalenii_PYR-1 MKIMAINLEMPKKLQAVIEKGHQGAAEMIRPISRKYDLKEHAYPVELDTL
Mflv_4456|M.gilvum_PYR-GCK ---MAINLELPKKLRAVIEKGHQGAAEILRPISRKYDEKEHAYPVELDTV
TH_3136|M.thermoresistible__bu ---MAINLELPKKLQAVVQKGHQGAAEMLRPISRKYDLQEHAYPVELDTL
MAB_3487|M.abscessus_ATCC_1997 ---MAINLELPKKLQAIVEKGHQGAAELVRPISRKYDLAEHAYPVELETL
Mb3164|M.bovis_AF2122/97 ---MAINLELPRKLQAIIVKTHQGAAEMMRPIARKYDLKEHAYPVELDTL
Rv3140|M.tuberculosis_H37Rv ---MAINLELPRKLQAIIVKTHQGAAEMMRPIARKYDLKEHAYPVELDTL
MMAR_1509|M.marinum_M ---MAINLEIPRKLQAIITKTHQGTAELIRPIARKYDLKEHAYPVELDTL
MUL_2432|M.ulcerans_Agy99 ---MAINLEIPRKLQAIITKTHQGTAELIRPIARKYDLKEHAYPVELDTL
MLBr_00660|M.leprae_Br4923 ---MAINLELSRKLQAVIVKTHQGAAELMRPIARKYDLKEHTYPVELDTL
MAV_4019|M.avium_104 ---MAINLELPRKMQAVTTKTHQGAAELMRPIARKYDLKEHAYPVELDTL
******:.:*::*: * ***:**::***:**** **:*****:*:
MSMEG_2080|M.smegmatis_MC2_155 ANLFEGISSAKTISFAGAEAFRDDANGSG---KDATVNGANMSALLNALE
Mvan_1906|M.vanbaalenii_PYR-1 ADIFDGITAAKTISFAGTDAFVG-ARDDG---QKSNVNGANMSALLNALE
Mflv_4456|M.gilvum_PYR-GCK ATLFEGISEAKTISFAGAEAFRDGDDD-----PKGNINGGNMSAVLNIIE
TH_3136|M.thermoresistible__bu ADLFEGISEAKTISFAGAEAFAEAGKNGDKAGKKANVNGGNMSAVLNVIE
MAB_3487|M.abscessus_ATCC_1997 ETLFAGVTAAGTFAFAGAEAFRAEENT-----AKQNVNGANMSALLNALE
Mb3164|M.bovis_AF2122/97 INLFEG--AAESFNFAGAHSLRDEDEGKD-----ENHNGANMAAVVQTME
Rv3140|M.tuberculosis_H37Rv INLFEG--AAESFNFAGAHSLRDEDEGKD-----ENHNGANMAAVVQTME
MMAR_1509|M.marinum_M ITLFEG--ASESYNFAGADALRGSEDSKD-----ENHNGANMAALVQTME
MUL_2432|M.ulcerans_Agy99 ITLFEG--ASESYNFAGADALRGSEDSKD-----ENHNGANMAALVQTME
MLBr_00660|M.leprae_Br4923 FNLFAG--AAESFAFAGADALGD-EDNKD-----ENHNGANMAALLQTLE
MAV_4019|M.avium_104 ISLFEG--ASESIEFAGANALGGEDEERD-----KNHNGANMAALLQTLE
:* * : : ***:.:: . **.**:*::: :*
MSMEG_2080|M.smegmatis_MC2_155 ISWGDVALLLSVPRQGLGNAAISSVATPEQLERLGKDVWAAMAITEPGFG
Mvan_1906|M.vanbaalenii_PYR-1 ISWGDVALLLSVPRQGLGNAAISSVATPEQLERLGKGVWAAMAITEPSFG
Mflv_4456|M.gilvum_PYR-GCK VSWGDVALVLSIPYQGLGNAAISGVATDEQLERLGK-VWAAMAITEPGFG
TH_3136|M.thermoresistible__bu ISWGDVALMLSVPRQGLGNAAISSVATPEQLERLGKDVWAAMAITEPGFG
MAB_3487|M.abscessus_ATCC_1997 MSWGDVGLLLTVPYQGLGNAAVSSVTSKEDGDRFGK-IWAAMAITEPGFG
Mb3164|M.bovis_AF2122/97 ASWGDVAMMLSLPYQGLGNAAISAVATDEQLERLGK-VWAAMAITEPEFG
Rv3140|M.tuberculosis_H37Rv ASWGDVAMMLSLPYQGLGNAAISAVATDEQLERLGK-VWAAMAITEPEFG
MMAR_1509|M.marinum_M ASWADVAMMLSIPYQGLGNAAISAVATDEQLERLGK-VWAAMAITEPAFG
MUL_2432|M.ulcerans_Agy99 ASWADVAMMLSIPYQGLGNAAISAVATDEQLERLGK-VWAAMAITEPAFG
MLBr_00660|M.leprae_Br4923 ACWGDVAMLLSIPYQGLGNAAISAVATNKQLERLGK-VWAAMAITEPGFG
MAV_4019|M.avium_104 ASWGDLAMMLSVPYQGLGNAAISAVANDEQLQRLGK-VWAAMAITEPGFG
.*.*:.::*::* *******:*.*:. :: :*:** :********* **
MSMEG_2080|M.smegmatis_MC2_155 SDSAAVSTTAVLDGDEYVINGEKIFVTAGSRATHIVVWATLDKSLGRAAI
Mvan_1906|M.vanbaalenii_PYR-1 SDSAAVSTTAVLDGDEYVINGEKIFVTAGSRASHIVVWATLDKSKGRAAI
Mflv_4456|M.gilvum_PYR-GCK SDSAAVSTTAKLDGDEYVINGEKIYVTAGSRATHIVVWASLDKSKGRAAI
TH_3136|M.thermoresistible__bu SDSAAVSTTATLDGDEYVINGEKIFVTAGSRASHIVVWATLDKSKGRAAI
MAB_3487|M.abscessus_ATCC_1997 SDSAAVSTTAKLDGDEYVINGEKIFVTAGSRATHIVVWATLDKTKGRPAI
Mb3164|M.bovis_AF2122/97 SDSAAVSTTATLDGDEYVINGEKIFVTAGSRATHIVVWATLDKSLGRPAI
Rv3140|M.tuberculosis_H37Rv SDSAAVSTTATLDGDEYVINGEKIFVTAGSRATHIVVWATLDKSLGRPAI
MMAR_1509|M.marinum_M SDSAAVSTTATLDGDEYVINGEKIFVTAGSRATHIVVWATLDKSKGRAAI
MUL_2432|M.ulcerans_Agy99 SDSAAVSTTATLDGDEYVINGEKIFVTAGSRATHIVVWATLDKSKGRAAI
MLBr_00660|M.leprae_Br4923 SDSAAVSTTATLDGDEYVINGEKIFVTAGSRATHIVVWATLDKSLGHAAI
MAV_4019|M.avium_104 SDSAAVSTTATLDGDEYVINGEKIFVTAGSRATHIVVWATLDKSKGRPAI
********** *************:*******:******:***: *:.**
MSMEG_2080|M.smegmatis_MC2_155 KSFIVPREHPGVTVERLERKLGIKASDTAVIRFENVRIPKDNLLGDPEIH
Mvan_1906|M.vanbaalenii_PYR-1 KSFIVPREHPGVTVERLEHKLGIKASDTAVIRFDNARIPKDNLLGDPEIH
Mflv_4456|M.gilvum_PYR-GCK KSFIVPREHPGVTVERLEHKLGIKASDTAAIRFDNVRIPRENLLGSPDIN
TH_3136|M.thermoresistible__bu KSFIVPRDHPGVTVERLEHKLGIKASDTATIRFDNVRIPKENLLGDPEIH
MAB_3487|M.abscessus_ATCC_1997 KSFLVPREHPGVTIERLEEKLGIKASDTAVIRFDNARIPKDYILSDPEIN
Mb3164|M.bovis_AF2122/97 KSFIVPREHPGVTVERLEHKLGIKGSDTAVIRFDNARIPKGNLLGNPEIE
Rv3140|M.tuberculosis_H37Rv KSFIVPREHPGVTVERLEHKLGIKGSDTAVIRFDNARIPKGNLLGNPEIE
MMAR_1509|M.marinum_M KSFIVPREHPGVIVERLEHKLGIKGSDTAVIRFDNARIPKDNLLGKPEIE
MUL_2432|M.ulcerans_Agy99 KSFIVAREHPGVIVERLEHKLGIKGSDTAVIRFDNARIPKDNLLGKPEIE
MLBr_00660|M.leprae_Br4923 KSFIVPREHPGVTVERLEYKLGIRGSDTAAIRFDNVRIPKDNLLGNPEIE
MAV_4019|M.avium_104 KSFIVPRDHPGVTVERLEKKLGIKGSDTAAIRFDNVRIPKENLLGNPEIE
***:*.*:**** :**** ****:.****.***:*.***: :*..*:*.
MSMEG_2080|M.smegmatis_MC2_155 VEKGFAGVMETFDNTRPIVAAMAVGIARAALEDLRAILTDAGVEISYDKP
Mvan_1906|M.vanbaalenii_PYR-1 VEKGFAGVMETFDNTRPIVAAMAVGVARAALEDLRKILTDAGIEISYDKP
Mflv_4456|M.gilvum_PYR-GCK VEKGFAGVMETFDNTRPVVAGMAVGVARAALEELRKILTDAGIEISYDKP
TH_3136|M.thermoresistible__bu VEKGFAGVMETFDNTRPIVAGMAVGIARAALEELRKILTDAGVEISYDKP
MAB_3487|M.abscessus_ATCC_1997 TEKGFGGVMSTFDNTRPIVAAMAVGVSRASLEELRKILEDAGIEISYDRP
Mb3164|M.bovis_AF2122/97 VGKGFAGVMETFDNTRPIVAAMAVGIGRAALEEIRSVLTGAGVEISYDKP
Rv3140|M.tuberculosis_H37Rv VGKGFAGVMETFDNTRPIVAAMAVGIGRAALEEIRSVLTGAGVEISYDKP
MMAR_1509|M.marinum_M VGKGFSGVMETFDNTRPIVAAMAVGIGRAALEEIRKILTDAGIDIDYDRP
MUL_2432|M.ulcerans_Agy99 MGKGFSGVMETFDNTRPIVAAMAVGIGRAALEEIRKILTDAGIDIDYDRP
MLBr_00660|M.leprae_Br4923 VGKGFSGVMETFDNTRPIVAAMAVGVGRAALEEIRKILTDAGIEICYDKP
MAV_4019|M.avium_104 EGKGFSGVMETFDNTRPIVAAMAVGVARAALEEIRKILTDAGVEISYDKP
***.***.*******:**.****:.**:**::* :* .**::* **:*
MSMEG_2080|M.smegmatis_MC2_155 AHAQSAAAAEFLRMEADWEAGYLLTVRSAWQADNNIPNSKEASMGKAKAA
Mvan_1906|M.vanbaalenii_PYR-1 AHAQSAAAAEFLRMEAEWESGYLLTVRSAWQADNRIPNSKEASMGKAKAA
Mflv_4456|M.gilvum_PYR-GCK AHAQSAAAAEFLRMEADWESGYLLTLRSAWQADNRIPNSKEASMGKAKAA
TH_3136|M.thermoresistible__bu AHAQSAPAAEFLRMEADWEAGYLLTLRAAWQADNGIPNSKEASMSKAKAA
MAB_3487|M.abscessus_ATCC_1997 AHAQHAAAAEFLRMEADWESSYLLTLRSAWQADNNVPNSMEASMAKAKAG
Mb3164|M.bovis_AF2122/97 SHTQSAAAAEFLRMEADWEASYLLSLRAAWQADNNIPNSKEASMSKAKAG
Rv3140|M.tuberculosis_H37Rv SHTQSAAAAEFLRMEADWEASYLLSLRAAWQADNNIPNSKEASMSKAKAG
MMAR_1509|M.marinum_M AYTQSAAAAEFLRMEADWEASYLLSLRAAWQADNKIPNSKEASISKAKAG
MUL_2432|M.ulcerans_Agy99 AYTQSAAAAEFLRMEADWEASYLLSLRAAWQADNKIPNSKEASISKAKAG
MLBr_00660|M.leprae_Br4923 SHSQNAAAAEFLRMEADWEASYLLSLRAAWQADNNIPNSKEASMSKAKAG
MAV_4019|M.avium_104 AHVQSAAAAEFLRMEADWESAYLLSLRAAWQADNKIPNSKEASMSKAKAG
:: * *.*********:**:.***::*:****** :*** ***:.****.
MSMEG_2080|M.smegmatis_MC2_155 RVASDITLKAVEMAGTTGYSEQTLLEKWARDSKILDIFEGTQQIQQLVVA
Mvan_1906|M.vanbaalenii_PYR-1 RVASDITLKAVEMAGTTGYSEETLLEKWARDSKILDIFEGTQQIQQLVVA
Mflv_4456|M.gilvum_PYR-GCK RVASDITLKTVELAGTTGYSEQALLEKWARDSKILDIFEGTQQIQQLVVA
TH_3136|M.thermoresistible__bu RVASDITLKAVELAGTVGYSEETLLEKWARDSKILDIFEGTQQIQLLVVA
MAB_3487|M.abscessus_ATCC_1997 RVGSDITLKAVEMAGTTGYSERTLLEKWGRDSKILDIFEGTQQIQQLVVA
Mb3164|M.bovis_AF2122/97 RMASDVTCKTVELAGTTGYSEQSLLEKWARDSKILDIFEGTQQIQQLVVA
Rv3140|M.tuberculosis_H37Rv RMASDVTCKTVELAGTTGYSEQSLLEKWARDSKILDIFEGTQQIQQLVVA
MMAR_1509|M.marinum_M RMASDVTLKVVELAGTTGYSEQLLLEKWGRDSKILDIFEGTQQIQQLVVA
MUL_2432|M.ulcerans_Agy99 RMASDVTLKVVELAGTTGYSEQLLLEKWGRDSKILDIFEGTQQIQQLVVA
MLBr_00660|M.leprae_Br4923 RMASDVTLKAVELAGTAGYSEKALLEKWARDSKILDIFEGTQQIQQLVVA
MAV_4019|M.avium_104 RMATDVTLKTVELAGTAGYSEQSLLEKWARDSKILDIFEGTQQIQQLVVA
*:.:*:* *.**:***.****. *****.**************** ****
MSMEG_2080|M.smegmatis_MC2_155 RRLLGLTSAELK
Mvan_1906|M.vanbaalenii_PYR-1 RRLLGLTSAQLK
Mflv_4456|M.gilvum_PYR-GCK RRLLGLSSAELK
TH_3136|M.thermoresistible__bu RRLLGLSSAELK
MAB_3487|M.abscessus_ATCC_1997 RRLLGLSSAELK
Mb3164|M.bovis_AF2122/97 RRLLGLSSSELK
Rv3140|M.tuberculosis_H37Rv RRLLGLSSSELK
MMAR_1509|M.marinum_M RRLLGMSSAELK
MUL_2432|M.ulcerans_Agy99 RRLLGMSSAELK
MLBr_00660|M.leprae_Br4923 RRLLGLSSSELK
MAV_4019|M.avium_104 RRLLGLSSAELK
*****::*::**