For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TFSLQLSDDVIEVRDWVHQFAAEVVRPAAAEWDEREETPWPVIQEAAKVGLYSPELFAQQAAEPTGIGML TVFEELFWGDAGIALSILGTGLAAAALAGNGTPEQLGQWLPEMFGTADEPKLGAFCSSEPDAGSDVGAIR TRARFDEATREWVLNGTKTWATNGGIANVHIVVASVYPELGSRGQATFIVPPGTHGLVQGQKFKKHGIRA SHTAEVVLDNVRLPEDMILGGREKFEERIARVKSGASAGGHAAMKTFERTRPTVGAMAVGVARAAYEYAL DYACQREQFGRKIGEFQAVAFKLADMKSRIDAARLLVWRAGWMARNNQQFDSAEGSMAKLVASETAVYVT DEAIQILGGNGYTRDYPVERMHRDAKIFTIFEGTSEIQRLVISRALTGLAIR*
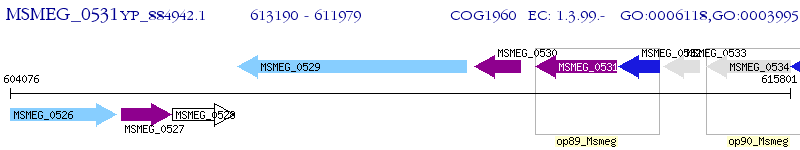
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0531 | - | - | 100% (403) | putative acyl-CoA dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_1821 | - | 3e-64 | 37.47% (395) | acyl-CoA dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_0108 | - | 2e-52 | 36.83% (391) | acyl-CoA dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3302c | fadE25 | 2e-62 | 36.96% (395) | acyl-CoA dehydrogenase FADE25 |
| M. gilvum PYR-GCK | Mflv_3829 | - | 0.0 | 80.40% (403) | acyl-CoA dehydrogenase domain-containing protein |
| M. tuberculosis H37Rv | Rv3274c | fadE25 | 2e-62 | 36.96% (395) | acyl-CoA dehydrogenase FADE25 |
| M. leprae Br4923 | MLBr_00737 | fadE25 | 6e-62 | 36.71% (395) | putative acyl-CoA dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3618c | - | 1e-60 | 35.95% (395) | acyl-CoA dehydrogenase FadE |
| M. marinum M | MMAR_0149 | fadE25_2 | 0.0 | 90.07% (403) | acyl-CoA dehydrogenase FadE25_2 |
| M. avium 104 | MAV_0145 | - | 0.0 | 93.80% (403) | putative acyl-CoA dehydrogenase |
| M. thermoresistible (build 8) | TH_3140 | - | 0.0 | 81.39% (403) | PUTATIVE acyl-CoA dehydrogenase domain protein |
| M. ulcerans Agy99 | MUL_4948 | fadE25_2 | 0.0 | 89.58% (403) | acyl-CoA dehydrogenase FadE25_2 |
| M. vanbaalenii PYR-1 | Mvan_2567 | - | 0.0 | 81.39% (403) | acyl-CoA dehydrogenase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_0531|M.smegmatis_MC2_155 -------MTFS-LQLSDDVIEVRDWVHQFAAEVVRPAAAEWDEREETPWP
MAV_0145|M.avium_104 -------MTFS-LQLSDDVIEVRDWVHQFAADVVRPAAAEWDEREETPWP
MMAR_0149|M.marinum_M -------MTFS-LELSDDVIEVRDWVHKFSAEVIRPAAAEWDEREETPWP
MUL_4948|M.ulcerans_Agy99 -------MTFS-LELSDDVIEVRDWVHKFSAEVIRPAAAEWDEREETPWP
Mflv_3829|M.gilvum_PYR-GCK -------MTFS-LELSSDLLDVQKWVHEFAADVVRPAAAEWDEREETPWP
Mvan_2567|M.vanbaalenii_PYR-1 -------MTFS-LELSSDLVDVQKWVHEFAADVVRPAAAEWDEREQTPWP
TH_3140|M.thermoresistible__bu -------MSFS-LELPPDLVHIQNWVHEFARDVIRPAAAEWDEREETPWP
Mb3302c|M.bovis_AF2122/97 MVGWAGNPSFDLFKLPEEHDEMRSAIRALAEKEIAPHAAEVDEKARFPEE
Rv3274c|M.tuberculosis_H37Rv MVGWAGNPSFDLFKLPEEHDEMRSAIRALAEKEIAPHAAEVDEKARFPEE
MLBr_00737|M.leprae_Br4923 MVGWSGNPLFDLFKLPEEHNELRATIRALAEKEIAPHAADVDQRARFPEE
MAB_3618c|M.abscessus_ATCC_199 ---MAGNPSFDLFKLGEEHDELRAAIRGLAEKEIAPHAKDVDEKARFPQE
*. ::* : .:: :: :: . : * * : *:: . *
MSMEG_0531|M.smegmatis_MC2_155 VIQEAAKVGLYSPELFAQQAAE-PTGIGMLTVFEELFWGDAGIALSILGT
MAV_0145|M.avium_104 VIQEAAKVGLYSPDLFAQQAAE-PTGLGMLTVFEELFWGDAGIALSIMGT
MMAR_0149|M.marinum_M VIQEAAKVGLYSPDFFAQQSVE-PSGVGILAAFEEMFWGDAGIAISIMGT
MUL_4948|M.ulcerans_Agy99 VIQEATKVGLYSPDFFAQQSVE-PSGVGILAAFEEMFWGDAGIAISIMGT
Mflv_3829|M.gilvum_PYR-GCK IIREAAKVGLYSMEFFAEQAAE-PSGLGMIVAFEEMFWGDAGIALAILGT
Mvan_2567|M.vanbaalenii_PYR-1 IIQEAAKVGLYSMEFFAEQAAE-PSGLGMIVAFEEMFWGDAGIALAILGT
TH_3140|M.thermoresistible__bu IIQEAAKVGIYSMEYFAEQAAE-PTGLGMPITFEEMFWGDAGIGLSILGT
Mb3302c|M.bovis_AF2122/97 ALVALNSSGFNAVHIPEEYGGQGADSVATCIVIEEVARVDASASLIPAVN
Rv3274c|M.tuberculosis_H37Rv ALVALNSSGFNAVHIPEEYGGQGADSVATCIVIEEVARVDASASLIPAVN
MLBr_00737|M.leprae_Br4923 ALAALNASGFNAIHVPEEYGGQGADSVAACIVIEEVARVDASASLIPAVN
MAB_3618c|M.abscessus_ATCC_199 ALDALVASGFNAVHVPEEYDGQGADSVAACIVIEEVARVCASSSLIPAVN
: *: : . : : . .:. .:**: *. .: .
MSMEG_0531|M.smegmatis_MC2_155 GLAAAALAGNGTPEQLGQWLPEMFGTADEPKLGAFCSSEPDAGSDVGAIR
MAV_0145|M.avium_104 GLAAAALAGNGTPEQLGRWLPEMFGTADEPKLGAFCSSEPDAGSDVGAIR
MMAR_0149|M.marinum_M GLAAAALAANGTPEQLGRWLPEMFGTPGDPKLGAFCSSEPDAGSDVGAIR
MUL_4948|M.ulcerans_Agy99 GLAAAALAANGTPEQLGRWLPEMFGTPGDPKLGAFCSSEPGAGSDVGAIR
Mflv_3829|M.gilvum_PYR-GCK GLAAASVAANGTPEQVGEWVPQMFGSVDDPKVAAFCSSEPNAGSDVGSIL
Mvan_2567|M.vanbaalenii_PYR-1 GLAAASLAANGTPEQVGEWVPQMFGSVDDPKVAAFCSSEPNAGSDVGSIL
TH_3140|M.thermoresistible__bu GLAAAALAGGGTLEQQAEYLPQMFGTVDDPKVAAFCASEPGAGSDVGAIR
Mb3302c|M.bovis_AF2122/97 KLGTMGLILRGSEELKKQVLPALAAEG---AMASYALSEREAGSDAASMR
Rv3274c|M.tuberculosis_H37Rv KLGTMGLILRGSEELKKQVLPALAAEG---AMASYALSEREAGSDAASMR
MLBr_00737|M.leprae_Br4923 KLGTMGLILRGSEELKKQVLPSLAAEG---AMASYALSEREAGSDAASMR
MAB_3618c|M.abscessus_ATCC_199 KLGTMGLILNGSDELKKQVLPAIASGE---AMASYALSEREAGSDAASMR
*.: .: *: * . :* : . :.::. ** ****..::
MSMEG_0531|M.smegmatis_MC2_155 TRARFDEATREWVLNGTKTWATNGGIANVHIVVASVYPELGSRGQATFIV
MAV_0145|M.avium_104 TRARYDEATGEWVLNGTKTWATNGGIANVHIVVASVYPELGARGQATFVV
MMAR_0149|M.marinum_M TRAHFDEAAGEWVLNGTKTWATNGGIANVHIVVVSVHPELGTRGQATFVI
MUL_4948|M.ulcerans_Agy99 TRAHFDEAAGEWVLNGTKTWATNGGIANVHIVVVSVHPELGTRGQATFVI
Mflv_3829|M.gilvum_PYR-GCK TRARYDEATDEWVLNGTKTWATNGGIANVHVVVASVYPELGTRGQVSFII
Mvan_2567|M.vanbaalenii_PYR-1 TRARYDEATDEWVLNGTKTWATNGGIANVHVVVASVHPELGTRGQASFII
TH_3140|M.thermoresistible__bu TRARYDKATDEWVLNGTKTWATNGGIAEVHIVMASVYPELGARGQAAFII
Mb3302c|M.bovis_AF2122/97 TRAKADG--DHWILNGAKCWITNGGKSTWYTVMAVTDPDRGANGISAFMV
Rv3274c|M.tuberculosis_H37Rv TRAKADG--DHWILNGAKCWITNGGKSTWYTVMAVTDPDRGANGISAFMV
MLBr_00737|M.leprae_Br4923 TRAKADG--DDWILNGFKCWITNGGKSTWYTVMAVTDPDKGANGISAFIV
MAB_3618c|M.abscessus_ATCC_199 TRAKADG--DDWIINGSKCWITNGGKSSWYTVMAVTDPEKKANGISAFMV
***: * .*::** * * **** : : *:. . *: :.* :*::
MSMEG_0531|M.smegmatis_MC2_155 PPGTHGLVQGQKFKKHGIRASHTAEVVLDNVRLPEDMILGGREKFEERIA
MAV_0145|M.avium_104 PAGVKGLAQGQKFKKHGIRASHTAEVVLDDVRLPADHILGGREKFEERIA
MMAR_0149|M.marinum_M PPGTKGLTQGQKFKKHGIRASHTAEVVLDNVRLPEDAILGGREKFEARIA
MUL_4948|M.ulcerans_Agy99 PPGTKGLTQGQKFKKHGIRASHTAEVVLDNVRLPEDAILGGREKFETRIA
Mflv_3829|M.gilvum_PYR-GCK PPGTKGLSQGQKFLKHGIRASHTAEVVLDDVRIPGRMIVGGKERFDARIA
Mvan_2567|M.vanbaalenii_PYR-1 PPGTPGLSQGQKFLKHGIRASHTAEVVLDDVRIPGRLIVGGKEKFDARIA
TH_3140|M.thermoresistible__bu PPGTKGFSQGQKFKKHGIRASHTAEVVLEDVRLPGRLIVGGREKFEERIA
Mb3302c|M.bovis_AF2122/97 HKDDEGFTVGPKERKLGIKGSPTTELYFENCRIPGDRIIGE---------
Rv3274c|M.tuberculosis_H37Rv HKDDEGFTVGPKERKLGIKGSPTTELYFENCRIPGDRIIGE---------
MLBr_00737|M.leprae_Br4923 HKDDEGFSIGPKEKKLGIKGSPTTELYFDKCRIPGDRIIGE---------
MAB_3618c|M.abscessus_ATCC_199 HKDDEGFTVGPLEHKLGIKGSPTAELYFENCRIPGDRIIGE---------
. *: * * **:.* *:*: ::. *:* *:*
MSMEG_0531|M.smegmatis_MC2_155 RVKSGASAGGHAAMKTFERTRPTVGAMAVGVARAAYEYALDYACQREQFG
MAV_0145|M.avium_104 RVKSGASARGQAALKTFERTRPTVGAMAVGVARAAYEYALEYACQREQFG
MMAR_0149|M.marinum_M RVKSGASARGQAAMKTFERTRPTVGAMAVGVARAAYEYALEYACQREQFG
MUL_4948|M.ulcerans_Agy99 RVKSGASARGQAAMKTFERTRPTVGAMAVGVARAAYEYALEYACQREQFG
Mflv_3829|M.gilvum_PYR-GCK KVREGKKAAGQAAMATFERTRPTVGAMAVGVARAAFEYARDYACEREQFG
Mvan_2567|M.vanbaalenii_PYR-1 KVREGKKAAGQAAMATFERTRPTVGAMAVGVARAAYEYALDYACEREQFG
TH_3140|M.thermoresistible__bu RVREGKSSKSQAAMATFERTRPTVGAMAVGVARAAYEYALEYALQREQFG
Mb3302c|M.bovis_AF2122/97 -----PGTGFKTALATLDHTRPTIGAQAVGIAQGALDAAIAYTKDRKQFG
Rv3274c|M.tuberculosis_H37Rv -----PGTGFKTALATLDHTRPTIGAQAVGIAQGALDAAIAYTKDRKQFG
MLBr_00737|M.leprae_Br4923 -----PGTGFKTALATLDHTRPTIGAQAVGIAQGALDAAIVYTKDRKQFG
MAB_3618c|M.abscessus_ATCC_199 -----PGTGFKTALQTLDHTRPTIGAQALGIAQGALDQAIAYTKDRKQFG
: ::*: *:::****:** *:*:*:.* : * *: :*:***
MSMEG_0531|M.smegmatis_MC2_155 RKIGEFQAVAFKLADMKSRIDAARLLVWRAGWMARNNQQFDSAEGSMAKL
MAV_0145|M.avium_104 RKIGEFQAVAFKLADMKCRIDAARMLVWRAGWMARNNQAFDAAEGSMAKL
MMAR_0149|M.marinum_M RKIGEFQAVAFKLADMKARIDAARLLVWRAGWMARNNHNFDSAEGSMAKL
MUL_4948|M.ulcerans_Agy99 RKIGEFQAVAFKLADMKARIDAARLLVWRAGWMARNNHNFDSAEGSMAKL
Mflv_3829|M.gilvum_PYR-GCK RKIGEFQGVAFKLADMKARIDAARLLVYRAGWMARNGKAFESAEGSMAKL
Mvan_2567|M.vanbaalenii_PYR-1 RKIGEFQAVAFKLADMKARIDAARLLVYRAGWMARNGKEFTAAEGSMAKL
TH_3140|M.thermoresistible__bu RKIAEFQAIAFKLADMKSRIDAARLLVWRAGWMARNGKSFDNAEGSMAKL
Mb3302c|M.bovis_AF2122/97 ESISTFQAVQFMLADMAMKVEAARLMVYSAAARAERGEPDLGFISAASKC
Rv3274c|M.tuberculosis_H37Rv ESISTFQAVQFMLADMAMKVEAARLMVYSAAARAERGEPDLGFISAASKC
MLBr_00737|M.leprae_Br4923 ESISTFQSIQFMLADMAMKVEAARLIVYAAAARAERGEPDLGFISAASKC
MAB_3618c|M.abscessus_ATCC_199 KSISDFQGVQFMLADMAMKVEAARLMVYTAAARAERGEKNLGFISSASKC
..*. **.: * **** :::***::*: *. *.... .: :*
MSMEG_0531|M.smegmatis_MC2_155 VASETAVYVTDEAIQILGGNGYTRDYPVERMHRDAKIFTIFEGTSEIQRL
MAV_0145|M.avium_104 VASETAVYVTDEAIQILGGNGYTRDYPVERMHRDAKIFTIFEGTSEIQRL
MMAR_0149|M.marinum_M VASETAVYVTDEAIQILGGNGYTRDYPVERMHRDAKIFTIFEGTSEIQRL
MUL_4948|M.ulcerans_Agy99 VASETAVYVTDEAIQILGGNGYTRDYPVERMHRDAKIFTIFEGTSEIQRL
Mflv_3829|M.gilvum_PYR-GCK VASETAVYVTDEAIQILGGNGYTREYPVERMHRDAKIFTIFEGTSEIQRL
Mvan_2567|M.vanbaalenii_PYR-1 VASETAVYVTDEAIQILGGNGYTREYPVERMHRDAKIFTIFEGTSEIQRL
TH_3140|M.thermoresistible__bu VASETATYVTDEAIQILGGNGYTRDYPVERMHRDAKIFTIFEGTSEIQRL
Mb3302c|M.bovis_AF2122/97 FASDVAMEVTTDAVQLFGGAGYTTDFPVERFMRDAKITQIYEGTNQIQRV
Rv3274c|M.tuberculosis_H37Rv FASDVAMEVTTDAVQLFGGAGYTTDFPVERFMRDAKITQIYEGTNQIQRV
MLBr_00737|M.leprae_Br4923 FASDIAMEVTTDAVQLFGGAGYTSDFPVERFMRDAKITQIYEGTNQIQRV
MAB_3618c|M.abscessus_ATCC_199 FASDVAMEVTTDAVQLFGGAGYTTDFPVERMMRDAKITQIYEGTNQIQRV
.**: * ** :*:*::** *** ::****: ***** *:***.:***:
MSMEG_0531|M.smegmatis_MC2_155 VISRALTGLAIR
MAV_0145|M.avium_104 VISRALTGLPIR
MMAR_0149|M.marinum_M VISRALTGLPIR
MUL_4948|M.ulcerans_Agy99 VISRALTGLPIR
Mflv_3829|M.gilvum_PYR-GCK VMARAITGLPIR
Mvan_2567|M.vanbaalenii_PYR-1 VMARALTGLPIR
TH_3140|M.thermoresistible__bu VIARALTGLPIR
Mb3302c|M.bovis_AF2122/97 VMSRALLR----
Rv3274c|M.tuberculosis_H37Rv VMSRALLR----
MLBr_00737|M.leprae_Br4923 VMSRALLR----
MAB_3618c|M.abscessus_ATCC_199 VMSRALLTS---
*::**: