For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAPVQKVVIITGAGSGIGQATAQVLLDAGHHVVLAGRRADALRGVAQGRPNARVVPTDVTDPASVRALFA DAVATFGRVDVLFNNAGVFGPSASVAEIDDEQWERTWRTNVDGSVFCAREAARVMSEQLPRGGRIINNGS LSAHRPRPNSLAYTVTKHAISGLTASMLLDLRSVDICVTQIDIGNAATSMTTGFSHQTLQADGSLAAEPV FDVVHVARAVAHLVDLPLEVAVPSLTVMAREMPFYGRG
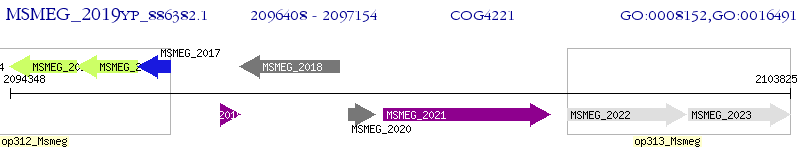
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2019 | - | - | 100% (248) | short-chain dehydrogenase/reductase SDR |
| M. smegmatis MC2 155 | MSMEG_4117 | - | 9e-25 | 36.36% (209) | 2-deoxy-D-gluconate 3-dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_2013 | - | 3e-24 | 37.02% (181) | oxidoreductase, short chain dehydrogenase/reductase family |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1079 | - | 7e-25 | 40.76% (184) | oxidoreductase |
| M. gilvum PYR-GCK | Mflv_1263 | - | 7e-23 | 40.12% (167) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv1050 | - | 8e-25 | 40.76% (184) | oxidoreductase |
| M. leprae Br4923 | MLBr_00862 | ephD | 8e-16 | 31.75% (189) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4060 | - | 5e-20 | 31.51% (238) | putative short chain dehydrogenase/reductase |
| M. marinum M | MMAR_0810 | - | 3e-21 | 31.40% (242) | short-chain type oxidoreductase |
| M. avium 104 | MAV_0602 | - | 2e-22 | 37.50% (184) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_4266 | - | 2e-22 | 38.92% (167) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_4554 | - | 1e-21 | 31.40% (242) | short-chain type oxidoreductase |
| M. vanbaalenii PYR-1 | Mvan_5156 | - | 1e-21 | 33.82% (204) | short chain dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1079|M.bovis_AF2122/97 --------------------------------------------------
Rv1050|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0810|M.marinum_M --------------------------------------------------
MUL_4554|M.ulcerans_Agy99 --------------------------------------------------
MAB_4060|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_1263|M.gilvum_PYR-GCK --------------------------------------------------
TH_4266|M.thermoresistible__bu --------------------------------------------------
MAV_0602|M.avium_104 --------------------------------------------------
Mvan_5156|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_2019|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 MPATQSIHKVPHQFVESPDGVRLAIYQSGQPEGPAIVLVHGFPDSHVLWD
Mb1079|M.bovis_AF2122/97 --------------------------------------------------
Rv1050|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0810|M.marinum_M --------------------------------------------------
MUL_4554|M.ulcerans_Agy99 --------------------------------------------------
MAB_4060|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_1263|M.gilvum_PYR-GCK --------------------------------------------------
TH_4266|M.thermoresistible__bu --------------------------------------------------
MAV_0602|M.avium_104 --------------------------------------------------
Mvan_5156|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_2019|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 GVVPLLAKRFRIVRYDNRGVGQSSVPKPVSAYTMTRFADDFAAVIDELSP
Mb1079|M.bovis_AF2122/97 --------------------------------------------------
Rv1050|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0810|M.marinum_M --------------------------------------------------
MUL_4554|M.ulcerans_Agy99 --------------------------------------------------
MAB_4060|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_1263|M.gilvum_PYR-GCK --------------------------------------------------
TH_4266|M.thermoresistible__bu --------------------------------------------------
MAV_0602|M.avium_104 --------------------------------------------------
Mvan_5156|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_2019|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 DRPVHVLAHDWGSVGVWEYLSRSGASDRVASFTSVSGPSQDQLVDYILSG
Mb1079|M.bovis_AF2122/97 --------------------------------------------------
Rv1050|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0810|M.marinum_M --------------------------------------------------
MUL_4554|M.ulcerans_Agy99 --------------------------------------------------
MAB_4060|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_1263|M.gilvum_PYR-GCK --------------------------------------------------
TH_4266|M.thermoresistible__bu --------------------------------------------------
MAV_0602|M.avium_104 --------------------------------------------------
Mvan_5156|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_2019|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 LRRPWRPRTFARAVSQLFRLTYMVLFSIPVLVPMVLWIALSSATIRRTMV
Mb1079|M.bovis_AF2122/97 --------------------------------------------------
Rv1050|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0810|M.marinum_M --------------------------------------------------
MUL_4554|M.ulcerans_Agy99 --------------------------------------------------
MAB_4060|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_1263|M.gilvum_PYR-GCK --------------------------------------------------
TH_4266|M.thermoresistible__bu --------------------------------------------------
MAV_0602|M.avium_104 --------------------------------------------------
Mvan_5156|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_2019|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 DNISTDQIHHSATLARDAAHSVKTYPANYFRAFSGRRRGKGVPVVNVPVQ
Mb1079|M.bovis_AF2122/97 --------------------------------------------------
Rv1050|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0810|M.marinum_M --------------------------------------------------
MUL_4554|M.ulcerans_Agy99 --------------------------------------------------
MAB_4060|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_1263|M.gilvum_PYR-GCK --------------------------------------------------
TH_4266|M.thermoresistible__bu --------------------------------------------------
MAV_0602|M.avium_104 --------------------------------------------------
Mvan_5156|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_2019|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 LIVNTMDRYVRPYGYDATARWVPRLWRRDIKAGHFSPMSHPQVMAAAVHD
Mb1079|M.bovis_AF2122/97 --------------------MARQRFRDQVVLITGAS-SGIGEATAKAFA
Rv1050|M.tuberculosis_H37Rv --------------------MARQRFRDQVVLITGAS-SGIGEATAKAFA
MMAR_0810|M.marinum_M --------------------MTTTDAGNRVAVVTGAS-SGIGEATARTLA
MUL_4554|M.ulcerans_Agy99 --------------------MTTTDAGNRVAVVTGAS-SGIGEATARTLA
MAB_4060|M.abscessus_ATCC_1997 --------------------MTNPTFDGRVAVVTGAS-SGIGAATAKSLA
Mflv_1263|M.gilvum_PYR-GCK -------------------MSDRFSMEGRVAVITGGG-TGIGRASALVLA
TH_4266|M.thermoresistible__bu -------------------VDDRFSMAGRVAVITGGG-TGIGRASALVLA
MAV_0602|M.avium_104 ----------MSLSEAPKEIAGHGLLEGKVVIVTAAAGTGIGSATAKRAL
Mvan_5156|M.vanbaalenii_PYR-1 ------------------MGLYGDQFKEKVAIVTGAG-GGIGQAYAEALA
MSMEG_2019|M.smegmatis_MC2_155 -----------------------MAPVQKVVIITGAG-SGIGQATAQVLL
MLBr_00862|M.leprae_Br4923 LADLVDGKQPSRALLRAQVGRSRDTFGDTLVSVTGAG-SGIGRETALALA
:. :*... *** *
Mb1079|M.bovis_AF2122/97 REGAVVALAARREGALRRVAREIEAAGG-RAMVAPLDVSSSESVRAMVAD
Rv1050|M.tuberculosis_H37Rv REGAVVALAARREGALRRVAREIEAAGG-RAMVAPLDVSSSESVRAMVAD
MMAR_0810|M.marinum_M AQGFHVVAVARRA---DRITALTDQIGG-TPIVADVTDG------AAVAA
MUL_4554|M.ulcerans_Agy99 AQGFHVVAVARRA---DRITALTDQIGG-TPIVADVTDG------AAVAA
MAB_4060|M.abscessus_ATCC_1997 ALGFHVVVAARRE---DRVKSLAGEIGG-TAFVLDVTDA------DSVAA
Mflv_1263|M.gilvum_PYR-GCK EHGADVVLAGRRAEPLKETAAEVEALGR-RALAVPTDVTDADACQALVDT
TH_4266|M.thermoresistible__bu EYGADIVLAGRRPEPLKKTAAEIEALGR-RALAVPTDVTAPDECRALVDA
MAV_0602|M.avium_104 AEGADVVISDHHERRLGETADQLAALGQGRVESVLCDVTSTAQVDALFAS
Mvan_5156|M.vanbaalenii_PYR-1 REGAAVVVADINTEGAQKVADGIKGEGG-NALAVRVDVSDPESAKGMAAQ
MSMEG_2019|M.smegmatis_MC2_155 DAGHHVVLAGRRADALRGVAQGRP-----NARVVPTDVTDPASVRALFAD
MLBr_00862|M.leprae_Br4923 RRGAEVVVSDIDEAAAKNTAAQIVASGG-VAYAYALDVSDAAAVEAFAEQ
* :.
Mb1079|M.bovis_AF2122/97 VVGEFGRIDVVFNNAGVSLV--GPVDAETFLDDTREMLEIDYLGTVRVVR
Rv1050|M.tuberculosis_H37Rv VVGEFGRIDVVFNNAGVSLV--GPVDAETFLDDTREMLEIDYLGTVRVVR
MMAR_0810|M.marinum_M LADGLSRVDVLVNNAGGAKG--LEPVADADLEHWRWMWETNVLGTLRVTR
MUL_4554|M.ulcerans_Agy99 LADGLSRVDVLVNNAGGAKE--LEPVADADLEHWRWMWETNVLGTLRVTR
MAB_4060|M.abscessus_ATCC_1997 LTQSLDRVDVLVNNAGGAWG--LEPVLEADLEHWRWMWETNVVGTLQVTR
Mflv_1263|M.gilvum_PYR-GCK TLAEFGRLDVLLNNAGGGE---TKSLMKWTDDEWHHVLDLNLSSAWYLSR
TH_4266|M.thermoresistible__bu TVGEFGRLDVLVNNAGGGA---TKPLMKWTDDEWHEVLNLNLSSAWYLSR
MAV_0602|M.avium_104 AHARMGRIDVLVNNAGLGG---QTPVVDMTDDEWDRVLDVTLTSVFRATR
Mvan_5156|M.vanbaalenii_PYR-1 TLSEFGGIDYLVNNAAIFGGMKLDFLITVDWDYYKKFMSVNMDGALVCTR
MSMEG_2019|M.smegmatis_MC2_155 AVATFGRVDVLFNNAGVFGP--SASVAEIDDEQWERTWRTNVDGSVFCAR
MLBr_00862|M.leprae_Br4923 VSAKHGVPDIVVNNAGIGQAG---RFLDTPPEEFDRVLAVNLGGVVNCCR
. * :.***. : . *
Mb1079|M.bovis_AF2122/97 EVLPIMKQQ-RSG-RIMNMSSVVGRKAFARFAGYSSAMHAIAGFSDALRQ
Rv1050|M.tuberculosis_H37Rv EVLPIMKQQ-RSG-RIMNMSSVVGRKAFARFAGYSSAMHAIAGFSDALRQ
MMAR_0810|M.marinum_M ALLPKLIAS-GDG-LIVTVTSIAALEIYDGGAGYTAAKHAQGALHRTLRG
MUL_4554|M.ulcerans_Agy99 ALLPKLIAS-GDG-LIVTVTSIAALEIYDGGAGYTAAKHAQGALHRTLRG
MAB_4060|M.abscessus_ATCC_1997 ALLPKLIDS-GDG-LIVTVTSIAALDVYDGGSGYTSAKHAQAALHRTLRG
Mflv_1263|M.gilvum_PYR-GCK AAAKPMIAQ-GSG-AIVNISSGAGLLAMPQAPIYGAAKAGLQNLTGSMAA
TH_4266|M.thermoresistible__bu AAAKPMIDQ-GRG-AIVNISSGAGLLAMPQAPVYAAAKAGLNNLTGSMAA
MAV_0602|M.avium_104 AALRYFRDA-PHGGVIVNNASVLGWRAQHSQSHYAAAKAGVMALTRCSAI
Mvan_5156|M.vanbaalenii_PYR-1 AVYRKMAKR-GGG-AIVNQSSTAAWLYS---NFYGLAKVGINGLTQQLAT
MSMEG_2019|M.smegmatis_MC2_155 EAARVMSEQLPRGGRIINNGSLSAHRPRPNSLAYTVTKHAISGLTASMLL
MLBr_00862|M.leprae_Br4923 AFGQRLVER-GTGGHIVNVSSMAAYAPLQSLSAYCTSKAATYMFSDCLRA
: * *:. * . * : . :
Mb1079|M.bovis_AF2122/97 ELRGSGIAVSVIHPALTQTPLLANVDPADMPPPFRSLTPIPVHWVAAAVL
Rv1050|M.tuberculosis_H37Rv ELRGSGIAVSVIHPALTQTPLLANVDPADMPPPFRSLTPIPVHWVAAAVL
MMAR_0810|M.marinum_M ELLGLPVRLTEIAPGAVET------------------------EFSLVRF
MUL_4554|M.ulcerans_Agy99 ELLGLPVRLTEIAPGAVET------------------------EFSLVRF
MAB_4060|M.abscessus_ATCC_1997 ELLGKPVRLTEVAPGAVET------------------------EFSVVRF
Mflv_1263|M.gilvum_PYR-GCK AWTRKGVRVNCIACGAVRT--------------------------PGLEA
TH_4266|M.thermoresistible__bu AWTRKGVRVNCIACGAIRT--------------------------PGLEA
MAV_0602|M.avium_104 EAAEYDVRINAVSPSIARH--------------------------KFLEK
Mvan_5156|M.vanbaalenii_PYR-1 ELGGQNIRVNAIAPGPIDT--------------------------EANRT
MSMEG_2019|M.smegmatis_MC2_155 DLRSVDICVTQIDIGNAAT-----------------------------SM
MLBr_00862|M.leprae_Br4923 ELDAVGVGLTTICPGVIDTN---------------------IIQSTRIDT
: :. : .
Mb1079|M.bovis_AF2122/97 DGVARRRARVVVPFQPRLLMVGDAFSPRYGDRVVRLLESKIFGRLIGSYR
Rv1050|M.tuberculosis_H37Rv DGVARRRARVVVPFQPRLLMVGDAFSPRYGDRVVRLLESKIFGRLIGSYR
MMAR_0810|M.marinum_M DGDEQRADAVYS-----------GMTPLVAADIAEVIG------FVASRP
MUL_4554|M.ulcerans_Agy99 DGDEQRADAVYS-----------GMTPLVAADIAEVIG------FVASRP
MAB_4060|M.abscessus_ATCC_1997 DGDQDRADAVYR-----------GITPLVAEDVAEIIA------FVASRP
Mflv_1263|M.gilvum_PYR-GCK DAKRQGFDIDMIG------------QTNGSGRIAEPDEIGYGVLFFASDA
TH_4266|M.thermoresistible__bu DAKRQGFDIDMIG------------QTNGSGRIAEPDEIGYGVLFFASDA
MAV_0602|M.avium_104 TSSADLLDRLSAG------------EAFG--RAAEPWEVAATIAFLASDY
Mvan_5156|M.vanbaalenii_PYR-1 TTPQEMVADIVKG------------IPLS--RMGEPEDLVGMLLFLLSDQ
MSMEG_2019|M.smegmatis_MC2_155 TTGFSHQTLQADG-------------SLAAEPVFDVVHVARAVAHLVDLP
MLBr_00862|M.leprae_Br4923 PTAKQERVRDRRGQLDMMFRLRRYGPDKVADTILSAIKKNKPIRPVAPEA
.
Mb1079|M.bovis_AF2122/97 GSVYRHQPTESAKAQAAQPERGYSSAR
Rv1050|M.tuberculosis_H37Rv GSVYRHQPTESAKAQAAQPERGYSSAR
MMAR_0810|M.marinum_M PHVNLDQIIIRPRDQASATRRANHQV-
MUL_4554|M.ulcerans_Agy99 PHVNLDQIIIRPRDQASATRRANHQV-
MAB_4060|M.abscessus_ATCC_1997 PHVNLDQIVVKPRDQASSTRRATRQ--
Mflv_1263|M.gilvum_PYR-GCK SSYCSGQTLYMHGGPGPAGV-------
TH_4266|M.thermoresistible__bu SSYCSGQTLYMHGGPGPAGV-------
MAV_0602|M.avium_104 SSYLTGEVISVSSQHP-----------
Mvan_5156|M.vanbaalenii_PYR-1 AKWITGQIFNVDGGQIIRS--------
MSMEG_2019|M.smegmatis_MC2_155 LEVAVPSLTVMAREMPFYGRG------
MLBr_00862|M.leprae_Br4923 YALYGISRVLPQVLRSTARIRVI----
.