For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SHLRGRSFGLRRVVWIGIWISIFQQLNGINIIFYYSTTLWQSVGFSESDSLLSSVITSAIFVVVTLVAIA VVDKIGRKPLLIAGGVGMAAMLAVMGLCFAAAVEAGGAVTLPGSYGPIALVAANLFVVAFGVSWGPVTWV MLGEIFPNSYRGPALAVAVAVQWIANFVVTVTFPPFAALSLPLSYCFYSACALLSALFVLRFVRETKGRE LEEMDDIAD*
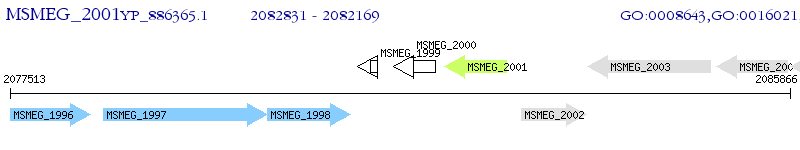
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2001 | - | - | 100% (220) | sugar transporter |
| M. smegmatis MC2 155 | MSMEG_4182 | - | 1e-63 | 57.00% (207) | arabinose-proton symporter |
| M. smegmatis MC2 155 | MSMEG_5559 | - | 5e-18 | 28.44% (211) | metabolite/sugar transport protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3364 | sugI | 6e-17 | 26.57% (207) | sugar-transport integral membrane protein SugI |
| M. gilvum PYR-GCK | Mflv_3055 | - | 3e-63 | 57.82% (211) | sugar transporter |
| M. tuberculosis H37Rv | Rv3331 | sugI | 1e-16 | 26.57% (207) | sugar-transport integral membrane protein SugI |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0692c | - | 8e-64 | 56.34% (213) | sugar transporter |
| M. marinum M | MMAR_1191 | sugI | 5e-18 | 27.05% (207) | sugar-transport integral membrane protein SugI |
| M. avium 104 | MAV_4306 | - | 2e-18 | 26.70% (206) | transporter, major facilitator superfamily protein |
| M. thermoresistible (build 8) | TH_2640 | - | 4e-64 | 57.01% (214) | PUTATIVE sugar transporter |
| M. ulcerans Agy99 | MUL_1444 | sugI | 2e-18 | 27.05% (207) | sugar-transport integral membrane protein SugI |
| M. vanbaalenii PYR-1 | Mvan_3472 | - | 3e-64 | 58.22% (213) | sugar transporter |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3055|M.gilvum_PYR-GCK ------------------------MAGQGGPAGESPELYEKGQDFSSGKT
Mvan_3472|M.vanbaalenii_PYR-1 ------------------------MAGQGGPVGDGPEIFEQGQEFSSGKT
TH_2640|M.thermoresistible__bu --------------------------------------------------
MAB_0692c|M.abscessus_ATCC_199 --------------------------------------MSEEDEFSSGHS
MSMEG_2001|M.smegmatis_MC2_155 --------------------------------------------------
Mb3364|M.bovis_AF2122/97 MTTLWQPHRNDYSPIPGRGVHARRGARRPRPRGGRAERPGTGQLTRSGRR
Rv3331|M.tuberculosis_H37Rv MTTLWQPHRNDYSPIPGRGVHARRGARRPRPRGGRAERPGTGQLTRSGRR
MMAR_1191|M.marinum_M -----------------------------------------------MQR
MUL_1444|M.ulcerans_Agy99 -----------------------------------------------MQR
MAV_4306|M.avium_104 -------------------------------------------MARGSRR
Mflv_3055|M.gilvum_PYR-GCK AIRIASVAALGGLLFGYDSAVINGAVASIQEDFGIGNYALGLAVASALLG
Mvan_3472|M.vanbaalenii_PYR-1 AVRIASVAALGGLLFGYDSAVINGAVDSIQEDFGIGNAELGFAVASALLG
TH_2640|M.thermoresistible__bu --------------------------------------------------
MAB_0692c|M.abscessus_ATCC_199 ALRIASVAALGGLLFGYDSAVINGAVQAIQDAFAIRDAELGFAVASALLG
MSMEG_2001|M.smegmatis_MC2_155 --------------------------------------------------
Mb3364|M.bovis_AF2122/97 ALLVGLTAASVGVLYGYDLSAIAGALLSLSEEFELTTREQELLTTTAVLG
Rv3331|M.tuberculosis_H37Rv ALLVGLTAASVGVLYGYDLSAIAGALLSLSEEFELTTREQELLTTTAVLG
MMAR_1191|M.marinum_M GILVALTAASVGLIYGYDLSIIAGAQLFITEEFGLSTHQQELLTTMVVIG
MUL_1444|M.ulcerans_Agy99 GILVALTAASVGLIYGYDLSIIAGAQLFITEEFGLSTHQQELLTTMVVIG
MAV_4306|M.avium_104 GLLVGLTAASVGVIYGYDLSIIAGAQLFVTEDFGLSTRQQELLTTMAVIG
Mflv_3055|M.gilvum_PYR-GCK AAVGALSAGRIADRIGRIAVMKIAAVLFLLSAFGTAFAPETITLVIGRIV
Mvan_3472|M.vanbaalenii_PYR-1 AAAGAMTAGRIADRIGRIAVMKIAAVLFLVSAFGTGFAHEVWAVVLFRIV
TH_2640|M.thermoresistible__bu --------------------------------------------------
MAB_0692c|M.abscessus_ATCC_199 AAVGAMTAGRVADRIGRVAVMKIAAALFLLSAVGAGLAPNIELLVLFRVI
MSMEG_2001|M.smegmatis_MC2_155 --------------------------------------------------
Mb3364|M.bovis_AF2122/97 QIAGALGGGILANAIGRKKSVVLIVAGYAVFALLGATSVSVPMLVVARLL
Rv3331|M.tuberculosis_H37Rv QIAGALGGGILANAIGRKKSVVLIVAGYAVFALLGATSVSVPMLVVARLL
MMAR_1191|M.marinum_M QIVGALGAGVLANAIGRKKSVVMLLVAYTMFAVLGSLSVSLPMLLAARFL
MUL_1444|M.ulcerans_Agy99 QIVGALGAGVLANAIGRKKSVVMLLVAYTMFAVLGALSVSLPMLLAARFL
MAV_4306|M.avium_104 QIGGALFAGVLANAIGRRRSVLLILSGYAVFALLAAFSVGLPMLLTARLL
Mflv_3055|M.gilvum_PYR-GCK GGVGVGVASVIAPAYIAETSPPGIRGRLGSLQQLAIVSGIFASFAVNYLL
Mvan_3472|M.vanbaalenii_PYR-1 GGIGVGVASVIAPAYIAETSPPGIRGRLGSLQQLAIVSGIFASFAVNWLL
TH_2640|M.thermoresistible__bu ---------------------------------LAIVSGIFLSLLIDGIL
MAB_0692c|M.abscessus_ATCC_199 GGVGVGVASLIAPAYIAETSPSRIRGRLGSLQQLAIVTGIFLSLTIDWLL
MSMEG_2001|M.smegmatis_MC2_155 --------------------------------------------------
Mb3364|M.bovis_AF2122/97 LGVTIGLSVVVVPVYVAESAPAAVRGSLVTAYQLATLSGIVVGYLVGYLL
Rv3331|M.tuberculosis_H37Rv LGVTIGLSVVVVPVYVAESAPAAVRGSLVTAYQLATLSGIVVGYLVGYLL
MMAR_1191|M.marinum_M LGLAVGVSIVVVPVYVAESAPAAVRGSLLTVYQLTTVSGLIVGYLTGYLL
MUL_1444|M.ulcerans_Agy99 LGLAVGVSIVVVPVYVAESAPAAVRGSLLTVYQLTTVSGLIVGYLTGYLL
MAV_4306|M.avium_104 LGLTIGVTVVVVPVYVAESAPTAVRGALLTAYQLAIVSGLIVGYLSGYLL
Mflv_3055|M.gilvum_PYR-GCK QWLAGGPNEPLWLGMDAWRWMFLAMAVPAVVYGGLAFTIPESPRYLVATH
Mvan_3472|M.vanbaalenii_PYR-1 QWAAGGPNEPLWFGLDAWRWMFLAMALPAVLYGVLAFTIPESPRYLVASH
TH_2640|M.thermoresistible__bu AALAGGSREELWLNMEAWRWMFLMMAVPAVLYGALTFTIPESPRYLVATH
MAB_0692c|M.abscessus_ATCC_199 AHLAGSSRDELWLGLPAWRWMFLAMALPALVYGTLAFTIPESPRYLVATH
MSMEG_2001|M.smegmatis_MC2_155 --------------------------------------------------
Mb3364|M.bovis_AF2122/97 AGSHG------------WRAMFGLAAAPATLLLPLLWRMPDTARWYLLKG
Rv3331|M.tuberculosis_H37Rv AGSHG------------WRAMFGLAAAPATLLLPLLWRMPDTARWYLLKG
MMAR_1191|M.marinum_M AGTHS------------WRWMLGLATVPAMLLLPLLIRMPDTPRWYVMKG
MUL_1444|M.ulcerans_Agy99 AGTHS------------WRWMLGLATVPAMLLLPLLIRMPDTPRWYVMKG
MAV_4306|M.avium_104 AGTHS------------WRWMLGLACVPAVLLLPLVFRMPDTARWYLLKG
Mflv_3055|M.gilvum_PYR-GCK KIPEARRVLSMLLGQKNLEITITRIRETLEREDRP---SWRDLKKPTGGI
Mvan_3472|M.vanbaalenii_PYR-1 KIPEARRVLSMLLGKKNLEITITRIRETLEREDKP---SWRDMRKPTGGL
TH_2640|M.thermoresistible__bu RVPEARRVLSRLLGAKNLEITINRIERSLRAEKPP---SWSDLRKPTGGM
MAB_0692c|M.abscessus_ATCC_199 RIPEARTVLATLLGEKNLDITIGRIQETLDQSTAP---SWRDLRKPTGGL
MSMEG_2001|M.smegmatis_MC2_155 ---------------------------------------MSHLRGRSFGL
Mb3364|M.bovis_AF2122/97 RIADARSALRRIQPEADIDAELADMAAAVDERGG----GIGEMVRRP--Y
Rv3331|M.tuberculosis_H37Rv RIADARSALRRIQPEADIDAELADMAAAVDERGG----GIGEMVRRP--Y
MMAR_1191|M.marinum_M RIQEARAALLRVDPAADVEEELAEIGTALSEGSG----GVSEMLRPP--F
MUL_1444|M.ulcerans_Agy99 RIQEARAALLRVDPAADVEEELAEIGTALSEGSG----GVSEMLRPP--F
MAV_4306|M.avium_104 RVDDARRALLRVEPAARVDDELAEIGRAVSEEAASLPAMLAEMVRSP--Y
.: .
Mflv_3055|M.gilvum_PYR-GCK YGIVWVGLGLSIFQQFVGINVIFYYSNVLWQAVGFS-ADESAVYTVITSV
Mvan_3472|M.vanbaalenii_PYR-1 YGIVWVGLGLSIFQQFVGINVIFYYSNVLWQAVGFS-ADESAVYTVITSV
TH_2640|M.thermoresistible__bu YGIVWVGLGLSIFQQFVGINVIFYYSNVLWEAVGFD-ESQSFLITVITSV
MAB_0692c|M.abscessus_ATCC_199 HAIVWIGVALAVFQQLVGINVIFYYSNVLWQAVGFG-ESSSFTITVITSI
MSMEG_2001|M.smegmatis_MC2_155 RRVVWIGIWISIFQQLNGINIIFYYSTTLWQSVGFS-ESDSLLSSVITSA
Mb3364|M.bovis_AF2122/97 LRATLFVIALGFLVQITGINAIIYYSPRLFAAMGFAGYFAMLALPAMVQV
Rv3331|M.tuberculosis_H37Rv LRATLFVIALGFLVQITGINAIIYYSPRLFAAMGFAGYFAMLALPAMVQV
MMAR_1191|M.marinum_M LRATIFVITLGFLIQITGINAIIYYSPRIFEAMGFTGDFALLGLPALVQI
MUL_1444|M.ulcerans_Agy99 LRATIFVITLGFLIQITGINAIIYYSPRIFEAMGFTGDFALLGLPALVQI
MAV_4306|M.avium_104 RRATVFVVVLGFLIQITGINAIIYYSPRIFEAMGFTGNFALLALPALVQV
. . : :..: *: *** *:*** :: ::** ..:..
Mflv_3055|M.gilvum_PYR-GCK VNVLTTLIAIALIDKIGRKPLLLIGSVGMAVTLSTMAFIFANAELIENEA
Mvan_3472|M.vanbaalenii_PYR-1 INVLTTLIAIALIDKIGRKPLLLIGSSGMAVTLITMAVIFANATVGADGN
TH_2640|M.thermoresistible__bu TNIVTTLIAIALIDKIGRKPLLLIGSTGMAGTLGVMAVIFGTAPVIDGQP
MAB_0692c|M.abscessus_ATCC_199 TNIATTLVAIALIDRVGRKPLLLIGSAGMAATLGTMAVIFGSATMVDGKP
MSMEG_2001|M.smegmatis_MC2_155 IFVVVTLVAIAVVDKIGRKPLLIAGGVGMAAMLAVMGLCFAAAVEAGGAV
Mb3364|M.bovis_AF2122/97 AGLAAVCASLFLVDRLGRRPILLSGIATMITADAVLITVFANDSDGGTGL
Rv3331|M.tuberculosis_H37Rv AGLAAVCASLFLVDRLGRRPILLSGIATMITADAVLITVFANDSDGGTGL
MMAR_1191|M.marinum_M AGVAAVITSLLLVDRVGRRPILLSGIAIMIVADVALMAVFAR---GQGAA
MUL_1444|M.ulcerans_Agy99 AGVAAVITSLLLVDRVGRRPILLSGIAIMIVADVALMAVFAR---GQGAA
MAV_4306|M.avium_104 AGLVAVGTALLLVDRVGRRPILLCGTAMMIVADVVLVAVFGR---GPGGV
: .. :: ::*::**:*:*: * * .: *.
Mflv_3055|M.gilvum_PYR-GCK GEMVPSLPGASGVIALIAANLFVVAFGMSWGPVVWVLLGEMFPNRIRAAA
Mvan_3472|M.vanbaalenii_PYR-1 ----PSLPGASGVIALIAANLFVVAFGMSWGPVVWVLLGEMFPNRIRAAA
TH_2640|M.thermoresistible__bu -----QLGDVAGPVALVAANLFVVSFGMSWGPVVWVLLGEMFPNRIRAAA
MAB_0692c|M.abscessus_ATCC_199 -----HLGPVAGPVALVAANLFVVAFGMSWGPVVWVLLGEIFPNRIRAAA
MSMEG_2001|M.smegmatis_MC2_155 -----TLPGSYGPIALVAANLFVVAFGVSWGPVTWVMLGEIFPNSYRGPA
Mb3364|M.bovis_AF2122/97 ------------VLGFAGVLLFIIGFNFGFGSLVWVYAAESFPSRLRSMG
Rv3331|M.tuberculosis_H37Rv ------------VLGFAGVLLFIIGFNFGFGSLVWVYAAESFPSRLRSMG
MMAR_1191|M.marinum_M ------------ILGFAGILLFIIGYTMGFGSLGWVYASESFPTRLRSIG
MUL_1444|M.ulcerans_Agy99 ------------ILGFAGILLFIIGYTMGFGSLGWVYASESFPTRLRSIG
MAV_4306|M.avium_104 ------------IAGFAGVLLFIFGYTMGFGSLGWVYASESFPSRLRSIG
.: . **:..: ..:*.: ** .* **. *. .
Mflv_3055|M.gilvum_PYR-GCK LGLAAAGQWTANWVITVSFPELRNF--LGAAYGFYALCAVLSGLFVWKWV
Mvan_3472|M.vanbaalenii_PYR-1 LGLAAAGQWAANWLITVTFPGLREH--LGLAYGFYGLCAVLSGLFVWRWV
TH_2640|M.thermoresistible__bu LGLAAAGQWTANWVITVTFPGLREH--LGPAYGFYALCAVLSLLFVWRWV
MAB_0692c|M.abscessus_ATCC_199 MGLATAGNWAANWAVTVTFPALRDA--LGIAYGCYALCAVLSLLFVARWV
MSMEG_2001|M.smegmatis_MC2_155 LAVAVAVQWIANFVVTVTFPPFAALS-LPLSYCFYSACALLSALFVLRFV
Mb3364|M.bovis_AF2122/97 SSLMLTSTLTANAIVAAFSLTMLRVLGGAGVFAVFGTFAVVAFVVVYRFA
Rv3331|M.tuberculosis_H37Rv SSPMLTSTLTANAIVAAFSLTMLRVLGGAGVFAVFGTFAVVAFVVVYRFA
MMAR_1191|M.marinum_M SSTMLTANLIANAIVAGVFLTMLHSLGGSGAFAVFGVLALVAFGVVYRYA
MUL_1444|M.ulcerans_Agy99 SSTMLTANLIANAIVAAVFLTMLHSLGGSGAFAVFGVLALVAFGVVYRYA
MAV_4306|M.avium_104 SSTMLTSNLVANAIVAAVFLTLLHSLGGAGTFAVFAVLAVVAFAFVHRYA
. : ** :: : : :. *::: .* ::.
Mflv_3055|M.gilvum_PYR-GCK RETKGVSLEDMHGEILHGDKS-ATG-------
Mvan_3472|M.vanbaalenii_PYR-1 METKGVSLEDMHGEILHADKT-ATG-------
TH_2640|M.thermoresistible__bu AETKGRTLEDMQPDVPGHDRASAPG-------
MAB_0692c|M.abscessus_ATCC_199 EETKGRALEDMDSAIS----------------
MSMEG_2001|M.smegmatis_MC2_155 RETKGRELEEMD-DIAD---------------
Mb3364|M.bovis_AF2122/97 PETKGRKLEEIRHFWENGGRWPAERSPAADEP
Rv3331|M.tuberculosis_H37Rv PETKGRKLEEIRHFWENGGRWPAERSPAADEP
MMAR_1191|M.marinum_M PETKGRQLEEIRHFWENGGRWPEKLTEAP---
MUL_1444|M.ulcerans_Agy99 PETKGRQLEEIRHFWENGGRWPEKLTEAP---
MAV_4306|M.avium_104 PETKGRQLEDIRHFWENGGRWD----------
**** **::