For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TAPTATVPPLAPDTADAGLVATLRRHAQGRFVGPVATLVLFVAMFGAVLDRYQFASPAQVFLNLLVDNAY LIVLAVGMTFVILTGGIDLSVGSVVALSTVILAKTLSLGWPAPVALVTVLAVGPLLGLTMGLIIEYFDVQ PFVATLAGMFLARGMCYVVSVDSIPINDPVLRNLGLTYLYLYDDKFIRWPVIIALAVVVGAMYVLHLTRF GKTVYAVGGNRQSAQLMGLQAARTRVSVYVISGFCASLAGILLAVQKLSGYSLNGVGLELDAIAAAVIGG VILAGGVGFVAGALVGVLVLGTIETFVTAENLDSYWTRIMTGALLLAFVLVQRVLVRKPR*
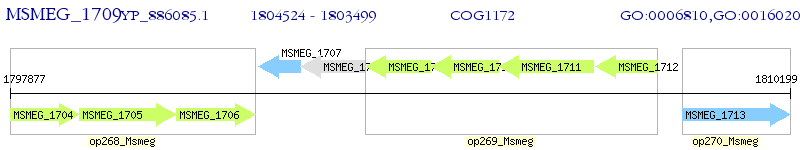
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1709 | - | - | 100% (341) | inner membrane ABC transporter permease protein YjfF |
| M. smegmatis MC2 155 | MSMEG_4171 | - | 9e-48 | 37.74% (318) | ribose transport system permease protein RbsC |
| M. smegmatis MC2 155 | MSMEG_3090 | - | 5e-43 | 36.36% (308) | ribose transport system permease protein RbsC |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2606 | - | 3e-23 | 26.40% (322) | inner-membrane translocator |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_00397 | - | 5e-38 | 33.12% (308) | putative transporter protein |
| M. abscessus ATCC 19977 | MAB_2625c | - | 6e-11 | 32.03% (153) | branched chain amino acid ABC transporter |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_1081 | - | 6e-10 | 32.14% (168) | ABC transporter branched chain amino acid transport permease |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_5291 | - | 6e-24 | 28.39% (391) | inner-membrane translocator |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_2625c|M.abscessus_ATCC_199 --------------------------------------------------
TH_1081|M.thermoresistible__bu --------------------------------------------------
MSMEG_1709|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_5291|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2606|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_00397|M.leprae_Br4923 MDHTTKCDAEQYFQAIVTSMADGVIVVDIDGRIESINPAATRILGLRAHD
MAB_2625c|M.abscessus_ATCC_199 --------------------------------------------------
TH_1081|M.thermoresistible__bu --------------------------------------------------
MSMEG_1709|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_5291|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2606|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_00397|M.leprae_Br4923 VVDMKHGHPFCFYDTDNQRVDLEREVMRVVRREVTTVSKVVGIDQHSGQR
MAB_2625c|M.abscessus_ATCC_199 --------------------------------------------------
TH_1081|M.thermoresistible__bu --------------------------------------------------
MSMEG_1709|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_5291|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2606|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_00397|M.leprae_Br4923 LWLSVNVSLLAYKAPPHSALVVSFSDISAHHLSIERLTYEATHDCLTGLA
MAB_2625c|M.abscessus_ATCC_199 --------------------------------------------------
TH_1081|M.thermoresistible__bu --------------------------------------------------
MSMEG_1709|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_5291|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2606|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_00397|M.leprae_Br4923 NRRFAEDQITKSLQHDERSRLAAVLLLDLDDFKVINDSLGHDVGDAVLQT
MAB_2625c|M.abscessus_ATCC_199 --------------------------------------------------
TH_1081|M.thermoresistible__bu --------------------------------------------------
MSMEG_1709|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_5291|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2606|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_00397|M.leprae_Br4923 VAQRLRSAVRPDDVVARLGGDEFIVLLRGPLSDMNANDVAKRLHTTLSES
MAB_2625c|M.abscessus_ATCC_199 -------------------MISGWMAQTGVQASTISFDLHGLWSSIGQLT
TH_1081|M.thermoresistible__bu -------------------MTHQCVALYTCHAADITFNLQGLREGLLQLT
MSMEG_1709|M.smegmatis_MC2_155 -------------------MTAPTATVPPLAPDTADAGLVATLRRHAQGR
Mvan_5291|M.vanbaalenii_PYR-1 --------MTTVRSDSDVSLADADFSGDTRADETFGAAVRGYLQRVRGGD
Mflv_2606|M.gilvum_PYR-GCK -------------------MSTKEALDVGGHKVVRDERVKERNRLQRLLI
MLBr_00397|M.leprae_Br4923 LVVDQLTVPIGASVGILEVRPDDRRRAADILRDADSAMYAAKNKKQCAVT
MAB_2625c|M.abscessus_ATCC_199 IDGLAWGAIYALVAVGYTLVFGVLRLINFAHSEVFMLGMFGAYFALDVVL
TH_1081|M.thermoresistible__bu IDGLSWGAIYALVAVGYTLVYGVLKLINFAHAEVFMLGMFGSYFCLDVIL
MSMEG_1709|M.smegmatis_MC2_155 FVGP--VATLVLFVAMFGAVLDRYQFASPAQVFLNLLVDNAYLIVLAVGM
Mvan_5291|M.vanbaalenii_PYR-1 MGSLPAVLGLVVLFVVFGLANDRFLSA---LNLANLITQAGSICVLAMGL
Mflv_2606|M.gilvum_PYR-GCK RPEMGAGIGAIGIFVFFLVVAPPFREAS---SLATVLYASSTIGIMACGV
MLBr_00397|M.leprae_Br4923 PQQLVPFVALIALFVFFTAAAGAKFYAP--SNLLVILQQTVVLAIVGYGM
. . : . :: : :
MAB_2625c|M.abscessus_ATCC_199 GFSPGGNAYNKGVSLTVLYLGVAMLVAMA-VSATTALGLEAVAYRPLRRR
TH_1081|M.thermoresistible__bu GFTPRGNAYDLGIGLTVLYLGIAMLFAIA-VSGTAAAGLEFIAYRPLRRR
MSMEG_1709|M.smegmatis_MC2_155 TFVILTGGIDLSVGSVVALSTVILAKTLS-LGWPAPVALVTVLAVGP---
Mvan_5291|M.vanbaalenii_PYR-1 VFVLLLGDIDLSAGVAGGVSACAMALIVVNLSWPWWAALLVGIACGA---
Mflv_2606|M.gilvum_PYR-GCK AVLMIGGEFDLSAGVAVTFSSLAASMLAYNLHLNLWVGALLALVLALT--
MLBr_00397|M.leprae_Br4923 TFVIMAGSVDLSVGSIVALTGVTAALVAAQNQFAAIVTALLVGLAAG---
. . : . .
MAB_2625c|M.abscessus_ATCC_199 GARPLTFLITAIGMSFVIQEFVHWVLPKIAKGYGGSNAQQP--LMLVTPR
TH_1081|M.thermoresistible__bu NAARLSFLITAIGMSFVLQEFVHFILPKILDGYGGSNAQQP--IRLVTPE
MSMEG_1709|M.smegmatis_MC2_155 ---LLGLTMGLIIEYFDVQPFVATLAGMFLARGMCYVVSVD--SIPINDP
Mvan_5291|M.vanbaalenii_PYR-1 ---LIGLAIGLLRAKLGIPSFVVTLAFFLGLQGVTLKLIGEGGSVRVDDP
Mflv_2606|M.gilvum_PYR-GCK ----VGFFNGFLVMKTKIPSFLITLSTFFMLAGINLAVTKLVAGQVATQS
MLBr_00397|M.leprae_Br4923 ------MVNGIVFAYGKIPSFVSTLGMLQVCRGITLMISDSSAKPMPFHG
: : : *: :
MAB_2625c|M.abscessus_ATCC_199 PVWTFGPFDVSGHHIPFQA-------------------------------
TH_1081|M.thermoresistible__bu TQFTLG--DVR---------------------------------------
MSMEG_1709|M.smegmatis_MC2_155 VLRNLGLTYLYLYD------------------------------------
Mvan_5291|M.vanbaalenii_PYR-1 VIRGLTISNLPVAAGWAIAGVVVLGFAGLELHQHRRKIALRLSHSPLGVV
Mflv_2606|M.gilvum_PYR-GCK VSDMQGWDSAQKVFASSFT-------------------------------
MLBr_00397|M.leprae_Br4923 ILGAMG--------------------------------------------
MAB_2625c|M.abscessus_ATCC_199 ---------------------------TITNVTVVVVAAALVLAFVTDIA
TH_1081|M.thermoresistible__bu ----------------------------VSNVTLIIVFAALVLALITDVV
MSMEG_1709|M.smegmatis_MC2_155 -----------------------------DKFIRWPVIIALAVVVGAMYV
Mvan_5291|M.vanbaalenii_PYR-1 VARVGAVAAVVLGVTYVMSVNRSVSAVADIRGIPYVLPLMLVLLIAMTVV
Mflv_2606|M.gilvum_PYR-GCK ---------------------------LFGVGIRITVVWWLVFTAVATWV
MLBr_00397|M.leprae_Br4923 -------------------------------AMPWILIVCLFVTILAGIL
: * .
MAB_2625c|M.abscessus_ATCC_199 INHTKFGRGIRAVAQDSTTATLMGVSRERIIMATFVIGGLLAGAAALLYT
TH_1081|M.thermoresistible__bu INRTRLGRGIRAVAQDPDTATLMGVSRERVILTTFVIGGLLAGAAALLYT
MSMEG_1709|M.smegmatis_MC2_155 LHLTRFGKTVYAVGGNRQSAQLMGLQAARTRVSVYVISGFCASLAGILLA
Mvan_5291|M.vanbaalenii_PYR-1 LRRTSYGRHIYAVGGNAEAARRAGISVDRIRVSVFVVCSSLAALSGIIAA
Mflv_2606|M.gilvum_PYR-GCK LFKTKVGNWIFAVGGDADSARAVGVPVTKVKIGLFMFVGFCAWFVGMHLL
MLBr_00397|M.leprae_Br4923 FQFTMFGRWVKAIGGNERVATLAGVPTRGIKVAIFAICGLTAGLGGIVLA
: * *. : *:. : * *: : : . . * .:
MAB_2625c|M.abscessus_ATCC_199 LKVPQGIIYSGGFLLGIKAFSAAVLG------GIGNLRGALLGGLILGVM
TH_1081|M.thermoresistible__bu LKMPQAIIYSGGFLLGIKAFTAAVLG------GIGNLRGALLGGLLLGVM
MSMEG_1709|M.smegmatis_MC2_155 VQKLSGYSLNG-VGLELDAIAAAVIGGVILAGGVGFVAGALVGVLVLGTI
Mvan_5291|M.vanbaalenii_PYR-1 SYAGKVSASSGAGNTLLYAVGAAVIGGTSLFGGKGRAIDAVIGGVVVATI
Mflv_2606|M.gilvum_PYR-GCK FAFNTVQSGQG-IGNEFFYIIAAVIGGCLLTGGYGTAIGAAIGALIFGMT
MLBr_00397|M.leprae_Br4923 SRLGAG-TPTAATGFEIDVIAAVVIGGTPLTGGLGRLSGTLIGAIIISML
. : . *.*:* * * .: :* ::..
MAB_2625c|M.abscessus_ATCC_199 ENYGQAVFGTQWRDVVAFVLLVLVLMFRPTGILGESLGRAKA
TH_1081|M.thermoresistible__bu ENYGQAVFGTQWRDVVAFVLLVLVLFFRPTGILGESLGKARV
MSMEG_1709|M.smegmatis_MC2_155 ETFVTAENLDSYWTRIMTGALLLAFVLVQRVLVRKPR-----
Mvan_5291|M.vanbaalenii_PYR-1 ANGLGLLNQSSYINFLVTGGVLLLAASVDAISRRRRSSTGLG
Mflv_2606|M.gilvum_PYR-GCK NQGIVYAGWDPDWFKFFLGAMLLFAVIANNAFRSYAARK---
MLBr_00397|M.leprae_Br4923 SNGMVFMGVGNAASQIIKGIMLAAAVFVFLQRRKIGIIK---
. ::