For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSTATASRSVEDAVRRRSGDLIELSHSIHAEPELAFAEHRSSAKTQALVAEYGFEITAAAGGLDTAFRAS YGSGSLVIGICAEYDALPEIGHACGHNIIASSAVGTALALAEVADLLDLTVVLVGTPAEETGGGKVLLLD AGVFDDIAASVMLHPGPVDIAAARSLALTEVAVSYTGRESHAAVAPYLGVNAADAVTVAQVAIGLLRQQL APGQMVHGIVTNGGQAANIIPGHAEMRYTMRAVDMAGLHELENRMAGCFAAGALATGCEHEVSETAPAYA ELRPDAWLSETVRAEMVRLGREPLPESVEASVPLGSTDMGNVTHLMPGIHPVIGVDSGGASLHQPAFAAA AAGPSADKAVVEGAIMLARTVVQLAETPAERDRVLAAQSRRVAS
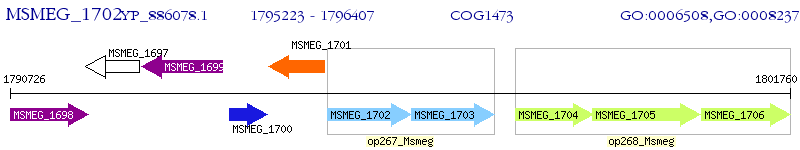
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1702 | - | - | 100% (394) | amidohydrolase |
| M. smegmatis MC2 155 | MSMEG_1679 | - | 7e-92 | 46.60% (382) | AmiB |
| M. smegmatis MC2 155 | MSMEG_3989 | - | 9e-15 | 25.67% (409) | peptidase M20D, amidohydrolase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3334c | amiB1 | 1e-169 | 74.81% (393) | amidase AmiB1 |
| M. gilvum PYR-GCK | Mflv_4827 | - | 1e-174 | 77.86% (393) | amidohydrolase |
| M. tuberculosis H37Rv | Rv3306c | amiB1 | 1e-169 | 74.81% (393) | amidohydrolase AmiB1 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3659c | - | 1e-151 | 68.99% (387) | putative peptidase/amidohydrolase |
| M. marinum M | MMAR_1220 | amiB1 | 1e-169 | 76.17% (386) | amidohydrolase AmiB1 |
| M. avium 104 | MAV_4282 | - | 1e-169 | 77.02% (383) | amidohydrolase |
| M. thermoresistible (build 8) | TH_1761 | - | 3e-84 | 73.30% (206) | PROBABLE AMIDOHYDROLASE AMIB1 (AMINOHYDROLASE) |
| M. ulcerans Agy99 | MUL_2672 | amiB1 | 1e-167 | 75.39% (386) | amidohydrolase AmiB1 |
| M. vanbaalenii PYR-1 | Mvan_1611 | - | 1e-175 | 78.12% (393) | amidohydrolase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3334c|M.bovis_AF2122/97 ---MPAAS---ASDRVEELVRRRGGELVELSHAIHAEPELAFAEHRSCAK
Rv3306c|M.tuberculosis_H37Rv ---MPAAS---ASDRVEELVRRRGGELVELSHAIHAEPELAFAEHRSCAK
MMAR_1220|M.marinum_M ---MPAL----VLDTVEELVRRRAGDLVELSHAIHAEPELAFAEHRSCAK
MUL_2672|M.ulcerans_Agy99 ---MPAL----VLDTVEELVRRRAGDLVELSHAIHAEPELAFAEHRSCAK
MAV_4282|M.avium_104 ---MPPVGSDSPLDSVEDVVRRRGADLVELSHAIHAEPELAFAEHRSCAK
MSMEG_1702|M.smegmatis_MC2_155 ---MSTAT---ASRSVEDAVRRRSGDLIELSHSIHAEPELAFAEHRSSAK
Mflv_4827|M.gilvum_PYR-GCK ---MSSAA---ASTSVEDAVNRRRSDLIELSHSIHAEPELAFAEHRSCAK
Mvan_1611|M.vanbaalenii_PYR-1 ---MPLAA---ASTCVEDAVRSRRADLVALSHAIHAEPELAFAEHRSCAK
TH_1761|M.thermoresistible__bu --------------------------------------------------
MAB_3659c|M.abscessus_ATCC_199 MPLTTGASSTAHAETASAVVDAAATDLVALSHDIHSEPELAFQEVRSALK
Mb3334c|M.bovis_AF2122/97 AQALVAERGFEITTAAGGLDTAFRADYGSGPLVVGVCAEYDALPGIGHAC
Rv3306c|M.tuberculosis_H37Rv AQALVAERGFEITTAAGGLDTAFRADYGSGPLVVGVCAEYDALPGIGHAC
MMAR_1220|M.marinum_M TQALVAERGFEITRAAAGLDTAFRADFGDGPLVVGICAEYDALPEIGHAC
MUL_2672|M.ulcerans_Agy99 TQALVAERGFEITRAAAGLDTAFRADFGDGPLVVGICAEYNALPEIGHAC
MAV_4282|M.avium_104 AQALVAERGFEITAAAGGLDTAFRADFGSGQLTIGVCAEYDALPEIGHAC
MSMEG_1702|M.smegmatis_MC2_155 TQALVAEYGFEITAAAGGLDTAFRASYGSGSLVIGICAEYDALPEIGHAC
Mflv_4827|M.gilvum_PYR-GCK TQALAAERGFEVSE-VAGLDTAFRAVYGSGPLVIGVCAEYDALPGIGHAC
Mvan_1611|M.vanbaalenii_PYR-1 TQALVAERGFEITAGVAGLDTAFRAVYGSGPLVVGVCAEYDALPEMGHAC
TH_1761|M.thermoresistible__bu --------------------------------------------------
MAB_3659c|M.abscessus_ATCC_199 TQRLLAERGFEIVTGQGGMDTAFRATYGHGPLVIGVCAEYDALPGIGHAC
Mb3334c|M.bovis_AF2122/97 GHNIIAASAVGTALALAEVADDLGLTVALLGTPAEESGGGKALMLQAGTF
Rv3306c|M.tuberculosis_H37Rv GHNIIAASAVGTALALAEVADDLGLTVALLGTPAEESGGGKALMLQAGTF
MMAR_1220|M.marinum_M GHNIIAASAVGTALALAEVADDLGLTVALLGTPAEESGGGKALMLDAGVF
MUL_2672|M.ulcerans_Agy99 GHNIIAASAVGTALALAEVADDLGLTVALLGTPAEESGGGKALMLDAGVF
MAV_4282|M.avium_104 GHNIIAASAVGTALALAEVADDLGLRVALVGTPAEEAGGGKALLLKAGVF
MSMEG_1702|M.smegmatis_MC2_155 GHNIIASSAVGTALALAEVADLLDLTVVLVGTPAEETGGGKVLLLDAGVF
Mflv_4827|M.gilvum_PYR-GCK GHNIIAASAVGTALALAEVADELGLTVVLLGTPAEELGGGKALMLDAGAF
Mvan_1611|M.vanbaalenii_PYR-1 GHNIIAASAVGTALALAEVADALGLTVVLLGTPAEELGGGKALMLQAGVF
TH_1761|M.thermoresistible__bu --------------------------------------------------
MAB_3659c|M.abscessus_ATCC_199 GHNIIAASAVGTGLGLAEVADALGLTVVVIGTPAEEFGGGKALLLKAGAF
Mb3334c|M.bovis_AF2122/97 DDVAVAVMVHPGPTDIAGARSLALSEVTVRYRGKESHAAVAPHLGVNAAD
Rv3306c|M.tuberculosis_H37Rv DDVAVAVMVHPGPTDIAGARSLALSEVTVRYRGKESHAAVAPHLGVNAAD
MMAR_1220|M.marinum_M DDITLALMVHPGPTDIAGARSLALSEVTVRYRGKESHAAVAPHLGVNAAD
MUL_2672|M.ulcerans_Agy99 DDITLALMVHPGPTDIAGARSLALSEVTVRYRGKESHAAVAPHLGVNAAD
MAV_4282|M.avium_104 DDISAAVMLHPGPADIAAARSLALSEAIVRYRGKESHAAVAPHLGVNALD
MSMEG_1702|M.smegmatis_MC2_155 DDIAASVMLHPGPVDIAAARSLALTEVAVSYTGRESHAAVAPYLGVNAAD
Mflv_4827|M.gilvum_PYR-GCK DDIAAAVMLHPGPLDIAAARSLALSEVTIGYTGRESHAAVAPHLGVNAGD
Mvan_1611|M.vanbaalenii_PYR-1 DDIAATVMLHPGPVDIAAARSLALSEMTIHYTGRESHAAVAPFLGVNAGD
TH_1761|M.thermoresistible__bu -------------------------------------------XGVNAAD
MAB_3659c|M.abscessus_ATCC_199 DDVAAAVMLHPGPIDIAGARSLALTDVMVTYHGKESHAAVAPQLGINAAD
*:** *
Mb3334c|M.bovis_AF2122/97 AVTVAQVAIGVLRQQLAPGQMVHGIVTDGGQAVNVIPGQARLQYAMRAVE
Rv3306c|M.tuberculosis_H37Rv AVTVAQVAIGVLRQQLAPGQMVHGIVTDGGQAVNVIPGQARLQYAMRAVE
MMAR_1220|M.marinum_M ALTIAQVAIGLLRQQLAPGQMTHGIVIDGGQAVNVIPGHAMMQYAMRAVE
MUL_2672|M.ulcerans_Agy99 ALTIAQVAIGLLRQQLAPGQMTHGIVIDGGQAVNVIPGHAMMQYAMRAVE
MAV_4282|M.avium_104 AVTVAQVAVGLLRQQLAPGQLVHGIVTDGGQAVNVIPGHASLQYAMRAVE
MSMEG_1702|M.smegmatis_MC2_155 AVTVAQVAIGLLRQQLAPGQMVHGIVTNGGQAANIIPGHAEMRYTMRAVD
Mflv_4827|M.gilvum_PYR-GCK AVTVTQVAIGLLRQQLAPGQMVHGIVTDGGQASNVIPAHTELRYTMRATN
Mvan_1611|M.vanbaalenii_PYR-1 AVTVAQVAIGLLRQQLAPGQMVHGIVTDGGQASNVIPGHTEMRYTMRAND
TH_1761|M.thermoresistible__bu AVTVAQVAIGLLRQHLEPGQQVHGIVTDGGQATNVIPAHAGMTYTMRAPH
MAB_3659c|M.abscessus_ATCC_199 AVTVAQVSIGLLRQHLSPGQLIHGIVTEGGEAANIIPGRAAMQYTMRAVE
*:*::**::*:***:* *** **** :**:* *:**.:: : *:*** .
Mb3334c|M.bovis_AF2122/97 SDSLRELQTRMFACFAAGALAAGCEYEIDEAAPAYAELKPDPWLADVCRE
Rv3306c|M.tuberculosis_H37Rv SDSLRELQTRMFACFAAGALAAGCEYEIDEAAPAYAELKPDPWLADVCRE
MMAR_1220|M.marinum_M SESLRDLEGRMFACFAAGALAAGCEYDIDAAAPAYAELKPDHWLVEVCRE
MUL_2672|M.ulcerans_Agy99 SESLRDLEGRMFACFAAGALAAGCGYDIDAAAPAYAELKPDHWLVEVCRE
MAV_4282|M.avium_104 SDSLRELEGRMYACFAAGAMAAGCEYEIDNPAPAYDELKPDAWLANIFRE
MSMEG_1702|M.smegmatis_MC2_155 MAGLHELENRMAGCFAAGALATGCEHEVSETAPAYAELRPDAWLSETVRA
Mflv_4827|M.gilvum_PYR-GCK SESLHELESRMAGCFASGAVATGCEHVVSETAPAYAELAPDPWLAERVRA
Mvan_1611|M.vanbaalenii_PYR-1 SESLRALESRMAGCFAAGAVATGCEHVVSDTAPAYAELAPDPWLSEVFRA
TH_1761|M.thermoresistible__bu AAALRALERRMADCFAAGALATGCSHTVTETSPPYDELAPDPWLAEVFRT
MAB_3659c|M.abscessus_ATCC_199 TASLRELEEKVYACFAAGAMATGCSHELVESSPPYLELTPDPWLVAVFRE
.*: *: :: ***:**:*:** : : .:*.* ** ** ** *
Mb3334c|M.bovis_AF2122/97 EMQRLGREPLLPALEAELPLGSTDMGNVTQVLPGIHPVIGLDAGAATVHQ
Rv3306c|M.tuberculosis_H37Rv EMQRLGREPLLPALEAELPLGSTDMGNVTQVLPGIHPVIGLDAGAATVHQ
MMAR_1220|M.marinum_M EMLRLGRTPVPASVEEGLPLGSTDMGNVTQVLPGIHPVIGVDAGGATVHQ
MUL_2672|M.ulcerans_Agy99 EMLRLGRTPVPASVEEGLPLGSTDMGNVTQVLPGIRPVIGVDAGGATVHQ
MAV_4282|M.avium_104 EMCRLGRQPVAPEYEAALPMGSTDMGNVTQALPGIHPVVGVESGGAMVHQ
MSMEG_1702|M.smegmatis_MC2_155 EMVRLGREPLPESVEASVPLGSTDMGNVTHLMPGIHPVIGVDSGGASLHQ
Mflv_4827|M.gilvum_PYR-GCK EMLRRGRTPVPAELEAEFPLGSTDMGNVTQVMPGIHPIVGIDAGGASIHQ
Mvan_1611|M.vanbaalenii_PYR-1 EMTRVGRTPVAAELEASLPLGSTDMGNVTQVMPGIHPIVGIDAGGASIHQ
TH_1761|M.thermoresistible__bu EMVRMGRAPVPAEVEATIPLGSTDMGNVTQVMPGIHPVVGIDSGGAAIHQ
MAB_3659c|M.abscessus_ATCC_199 EMESFGRTPVPAEFEATVPLGSTDMGNVTQVLPGIHPVIGIDSGGAVIHQ
** ** *: * .*:*********: :***:*::*:::*.* :**
Mb3334c|M.bovis_AF2122/97 RAFTVASAGASADRAVVDGAIMLARTVVRLAQTPDERDRVLAAQQRRAAR
Rv3306c|M.tuberculosis_H37Rv RAFTVASAGASADRAVVDGAIMLARTVVRLAQTPDERDRVLAAQQRRAAR
MMAR_1220|M.marinum_M RAFAAAAAGHSADQAVVDGAIMLARTAVALAENPDQRDRVLAAQQRRAAS
MUL_2672|M.ulcerans_Agy99 RAFAAAAAGHSADQAVVDGAIMLARTAVALAENPDQRDRVLAAQQRRAAS
MAV_4282|M.avium_104 RAFADAAAGPSADRAVIEGAIMLARTVVALAQTPAERDRVLAAHERRRGG
MSMEG_1702|M.smegmatis_MC2_155 PAFAAAAAGPSADKAVVEGAIMLARTVVQLAETPAERDRVLAAQSRRVAS
Mflv_4827|M.gilvum_PYR-GCK PAFTAAAVNASADAAVVDGAIMLARTVVALAESDVERDRVLDLWRKRAS-
Mvan_1611|M.vanbaalenii_PYR-1 PEFAAAAVSASADTAVVEGAIMLARTVVALAESGVERDRVLEQWQRRAS-
TH_1761|M.thermoresistible__bu PEFAAAAVSPSADRAVVEGAVMLARTVVRLAETPAERDRVLQRREARRTS
MAB_3659c|M.abscessus_ATCC_199 PDFEKAAREPGADRAVVEGAKMLARTVIRLAEDESERSRVLDRLARRTVK
* *: .** **::** *****.: **: :*.*** *
Mb3334c|M.bovis_AF2122/97 ---
Rv3306c|M.tuberculosis_H37Rv ---
MMAR_1220|M.marinum_M ---
MUL_2672|M.ulcerans_Agy99 ---
MAV_4282|M.avium_104 AHP
MSMEG_1702|M.smegmatis_MC2_155 ---
Mflv_4827|M.gilvum_PYR-GCK ---
Mvan_1611|M.vanbaalenii_PYR-1 ---
TH_1761|M.thermoresistible__bu ---
MAB_3659c|M.abscessus_ATCC_199 V--