For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAKNASSEADSSIIVDLSEAAMHMYSAAIDALPFPEDKKFHKRADIVLSGLRKLRSALTEAASAQRPSPA VAAALSGVRQRYDSLMAHAAAAPGSSLGQQIYVTRVAAKLSAEEVANGAGLRPGLLDELEAGATPTDDEA ARIRETIAALGGLPGADHGEHEGRHAGGDDSAEGASVIDFAQHDSQHEGGDGELTENAG
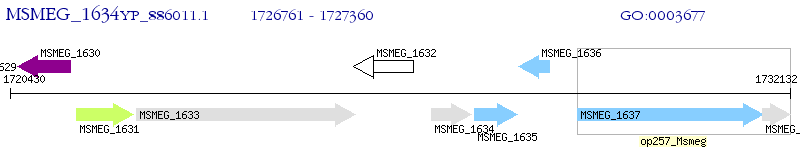
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1634 | - | - | 100% (199) | forkhead-associated protein |
| M. smegmatis MC2 155 | MSMEG_6518 | - | 7e-34 | 55.81% (129) | hypothetical protein MSMEG_6518 |
| M. smegmatis MC2 155 | MSMEG_0052 | - | 2e-14 | 34.65% (127) | hypothetical protein MSMEG_0052 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3893 | - | 1e-17 | 36.11% (144) | hypothetical protein Mb3893 |
| M. gilvum PYR-GCK | Mflv_0777 | - | 1e-18 | 34.23% (149) | hypothetical protein Mflv_0777 |
| M. tuberculosis H37Rv | Rv3863 | - | 1e-17 | 36.11% (144) | hypothetical protein Rv3863 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1298c | - | 2e-38 | 52.41% (145) | hypothetical protein MAB_1298c |
| M. marinum M | MMAR_5438 | - | 1e-17 | 38.10% (126) | putative regulatory protein |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_1779 | - | 5e-36 | 52.21% (136) | conserved hypothetical protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_0068 | - | 3e-17 | 33.56% (146) | hypothetical protein Mvan_0068 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_1634|M.smegmatis_MC2_155 --------------------------------------------------
TH_1779|M.thermoresistible__bu --------------------------------------------------
MAB_1298c|M.abscessus_ATCC_199 --------------------------------------------------
Mb3893|M.bovis_AF2122/97 --MAGERKVCPPSRLVPANKGSTQMSKAGSTVGPAPLVACSGGTSDVIEP
Rv3863|M.tuberculosis_H37Rv --MAGERKVCPPSRLVPANKGSTQMSKAGSTVGPAPLVACSGGTSDVIEP
MMAR_5438|M.marinum_M MLVSGTRALPPAS--VAASEESTYMPDAESTARPALRVARGGDSPGLVTP
Mflv_0777|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0068|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1634|M.smegmatis_MC2_155 --------------------------------------------------
TH_1779|M.thermoresistible__bu --------------------------------------------------
MAB_1298c|M.abscessus_ATCC_199 --------------------------------------------------
Mb3893|M.bovis_AF2122/97 RRGVAIIGHSCRVGTQIDDSRISQTHLRAVSDDGRWRIVGNIPRGMFVGG
Rv3863|M.tuberculosis_H37Rv RRGVAIIGHSCRVGTQIDDSRISQTHLRAVSDDGRWRIVGNIPRGMFVGG
MMAR_5438|M.marinum_M --GHRPVGRGALTNARLDDPVLSQTHVRAVSDGGQRRIVTNSPNGMFVDG
Mflv_0777|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0068|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1634|M.smegmatis_MC2_155 --------------------------------------------------
TH_1779|M.thermoresistible__bu --------------------------------------------------
MAB_1298c|M.abscessus_ATCC_199 --------------------------------------------------
Mb3893|M.bovis_AF2122/97 RRGSSVTVSDKTLIRFGDPPGGKALTFEVVRPSDSAAQHGRVQPSADLSD
Rv3863|M.tuberculosis_H37Rv RRGSSVTVSDKTLIRFGDPPGGKALTFEVVRPSDSAAQHGRVQPSADLSD
MMAR_5438|M.marinum_M TRKSSVAVSDKTIVRFGDPTGGKALTFEVVRPSNSPEEDSREQRPAEQSD
Mflv_0777|M.gilvum_PYR-GCK ------------------------------------------MTNAEPID
Mvan_0068|M.vanbaalenii_PYR-1 ------------------------------------------MTNAEPVD
MSMEG_1634|M.smegmatis_MC2_155 --------------------------------------------------
TH_1779|M.thermoresistible__bu --------------------------------------------------
MAB_1298c|M.abscessus_ATCC_199 --------------------------------------------------
Mb3893|M.bovis_AF2122/97 DPAHNAAPVAPDPGVVRAGAAAAARRRELDISQRSLAADGIINAGALIAF
Rv3863|M.tuberculosis_H37Rv DPAHNAAPVAPDPGVVRAGAAAAARRRELDISQRSLAADGIINAGALIAF
MMAR_5438|M.marinum_M SQTN-----EADPGVVRAGAAAAARRRELDISQRSLAADGIINAGALIAF
Mflv_0777|M.gilvum_PYR-GCK ESIG-----APDPGMARAGAAAAARRRELNISQRSLAAGGIINAGALIAF
Mvan_0068|M.vanbaalenii_PYR-1 QSTG-----APDPGMVRAGAAAAARRRELDISQRSLAAEGIINAGALIAF
MSMEG_1634|M.smegmatis_MC2_155 -------------------------------------------MAKNASS
TH_1779|M.thermoresistible__bu --------------------------------------------------
MAB_1298c|M.abscessus_ATCC_199 -------------------------------------------MPEVVPD
Mb3893|M.bovis_AF2122/97 EKGRSWPRERTRAKLEEVLQWPAGTIARIRRGEPTEPATNPDASPGLRPA
Rv3863|M.tuberculosis_H37Rv EKGRSWPRERTRAKLEEVLQWPAGTIARIRRGEPTEPATNPDASPGLRPA
MMAR_5438|M.marinum_M EKGRSWPRERTRAKLEEVLQWPAGTIARIRQGESVAPEPAPEASVEAPTS
Mflv_0777|M.gilvum_PYR-GCK EKGRSWPRERTRARLEEILQWPPGTIAALRRGEPVHPAPTP-EARPPASG
Mvan_0068|M.vanbaalenii_PYR-1 EKGRSWPRERTRARLEEILQWPPGTIARLRRGEPLRAAPPPPESRAPASN
MSMEG_1634|M.smegmatis_MC2_155 EADSSIIVDLSEAAMHMYSAAIDALPFPEDKKFHKRADIVLSGLRKLRSA
TH_1779|M.thermoresistible__bu ---------MADAAMHMYAAAIDALPDPADPEYPEQVGVVLAGLRKLQGA
MAB_1298c|M.abscessus_ATCC_199 DGEAPVIVSLVDAAVHMYSSAIDTLPDPSDPEYGERVAVVLSGLRKLESA
Mb3893|M.bovis_AF2122/97 DGPASLIAQAVTAAVDGCSLAIAALPATEDPEFTERAAPILADLRQLEAI
Rv3863|M.tuberculosis_H37Rv DGPASLIAQAVTAAVDGCSLAIAALPATEDPEFTERAAPILADLRQLEAI
MMAR_5438|M.marinum_M DGPASLIAQAVAAAVDTCSLAIAALPAPEEPDFTERAAPILADLRQLEGI
Mflv_0777|M.gilvum_PYR-GCK D-EVSLIAQAVITAVHAFGSTISSLPPADDAAFIPRVTAILADLRQLEAV
Mvan_0068|M.vanbaalenii_PYR-1 D-EVSLIAQAVTTAVSAFGSTVAALPPVDDAAFVPRVTAILADLRQLDAV
:*: . :: :** : : :. :*:.**:* .
MSMEG_1634|M.smegmatis_MC2_155 LTEAASAQRPSPAVAAALSGVRQRYDSLMAHAAAAPGSSLGQQIYVTRVA
TH_1779|M.thermoresistible__bu LAQAATRSRTNPSVIVALSAVRNRYDEVMAAAANGPNATLGQRLYIARQR
MAB_1298c|M.abscessus_ATCC_199 ISKAAGRSRVTPSVIVALSGVRHRYDDLMKAAANSPSATLGQRLYTARRR
Mb3893|M.bovis_AF2122/97 AVQATRISRITPELIKALGAVRRHHDELMRLGATAPGATLAQRLYAARRR
Rv3863|M.tuberculosis_H37Rv AVQATRISRITPELIKALGAVRRHHDELMRLGATAPGATLAQRLYAARRR
MMAR_5438|M.marinum_M AVQATRISRITPELIKALGAVRRYHDKLMTLSATAPGATLAQRLYAARRR
Mflv_0777|M.gilvum_PYR-GCK AARAARIGRVTPALIKALSTVRGLYDDLMLRAAGAPQAGLGHRLYAARRG
Mvan_0068|M.vanbaalenii_PYR-1 ASRAARIGRVTPALIKALSAVRGLYDDLMVRAARAPHATLGQRLYAARRG
.*: * .* : **. ** :*.:* .* .* : *.:::* :*
MSMEG_1634|M.smegmatis_MC2_155 AKLSAEEVANGAGLRPGLLDELEAGATPTDDEAARIRETIAALGGLPGAD
TH_1779|M.thermoresistible__bu AKLSIPEAANGIGLRPDIIAAVEAEEIPTEGEEIKIKDLIAALGG-----
MAB_1298c|M.abscessus_ATCC_199 ARLTAQETANGAGLKVGFLTAIESEEPVTEDEAAKIKDLIAALGG-----
Mb3893|M.bovis_AF2122/97 ANLSTLETAQAAGVAEEMIVGAEAEEELPAEATEAIEALIRQIN------
Rv3863|M.tuberculosis_H37Rv ANLSTLETAQAAGVAEEMIVGAEAEEELPAEATEAIEALIRQIN------
MMAR_5438|M.marinum_M ANLSTSETAQAAGVTEELIVRAEAEEALHAEAAEAIEALIRQIN------
Mflv_0777|M.gilvum_PYR-GCK ANLTVLETAQAAGVSERAIEQAEAGEPVTASEADAIETLVGQIA------
Mvan_0068|M.vanbaalenii_PYR-1 ANLTVLETAQAAGVSEDAVQQAEADEPLSPADIAAIETLISQIG------
*.*: *.*:. *: : *: *. : :
MSMEG_1634|M.smegmatis_MC2_155 HGEHEGRHAGGDDSAEGASVIDFAQHDSQHEGGDGELTENAG
TH_1779|M.thermoresistible__bu ------------------------------------------
MAB_1298c|M.abscessus_ATCC_199 ------------------------------------------
Mb3893|M.bovis_AF2122/97 ------------------------------------------
Rv3863|M.tuberculosis_H37Rv ------------------------------------------
MMAR_5438|M.marinum_M ------------------------------------------
Mflv_0777|M.gilvum_PYR-GCK ------------------------------------------
Mvan_0068|M.vanbaalenii_PYR-1 ------------------------------------------