For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
STPTMDDAAKALADPTAYADDARLHEALARLRAENPVAWVDQAPYRPFWAITKHADIMAIERANDLWLSA PRPLLATAEADDLGRSQQEMGIGLRTLIHMDDPHHRKVRAIGADWFRPKAMRELKVRVDELARIYVDKMR EIGPECDFVTDIAVNFPLYVILSLLGLPEEDFGRMHMLTQEMFGGDDDEYKRGTTVEEQMAVLTDFFNYF SALTNSRRENPTDDLASAIANGRVDGELMSDMDTLSYYVIVASAGHDTTKDAISGGLHALIENPGELARL KADPGLMGTAVEEMIRWSTPVKEFMRTAAEDTEVRGVPIAKGESVYLAYVSGNRDEEVFTDPFRFDVGRD PNKHLAFGYGVHFCLGAALARMEMNSLFSELLPRLDSIELAGEPELSATTFVGGLKHLPIRYSIR*
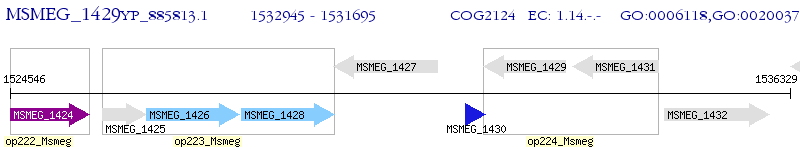
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1429 | - | - | 100% (416) | cytochrome P450-terp |
| M. smegmatis MC2 155 | MSMEG_1431 | - | e-174 | 71.39% (416) | cytochrome P450-terp |
| M. smegmatis MC2 155 | MSMEG_2257 | - | e-168 | 68.51% (416) | cytochrome P450-terp |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2289 | cyp124 | 1e-61 | 35.14% (407) | cytochrome P450 124 CYP124 |
| M. gilvum PYR-GCK | Mflv_5053 | - | 0.0 | 88.81% (420) | cytochrome P450 |
| M. tuberculosis H37Rv | Rv2266 | cyp124 | 2e-61 | 35.14% (407) | cytochrome P450 124 CYP124 |
| M. leprae Br4923 | MLBr_02088 | - | 8e-39 | 27.32% (410) | putative cytochrome p450 |
| M. abscessus ATCC 19977 | MAB_3825 | - | 1e-164 | 66.91% (411) | cytochrome P450 |
| M. marinum M | MMAR_3999 | cyp108B4 | 1e-167 | 69.16% (415) | cytochrome P450 108B4 Cyp108B4 |
| M. avium 104 | MAV_1609 | - | 1e-171 | 70.60% (415) | cytochrome P450 family protein |
| M. thermoresistible (build 8) | TH_4183 | - | 1e-178 | 73.33% (420) | PUTATIVE cytochrome P450 |
| M. ulcerans Agy99 | MUL_3863 | cyp108B4 | 1e-166 | 68.92% (415) | cytochrome P450 108B4 Cyp108B4 |
| M. vanbaalenii PYR-1 | Mvan_1301 | - | 0.0 | 87.38% (420) | cytochrome P450 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2289|M.bovis_AF2122/97 --------MGLNTAIATRVNGTPPPEVPIADIELGSLDFWALDDDVRDGA
Rv2266|M.tuberculosis_H37Rv --------MGLNTAIATRVNGTPPPEVPIADIELGSLDFWALDDDVRDGA
Mflv_5053|M.gilvum_PYR-GCK --------------MSTPTMDRDSQEAAKVLADP----KAYADDDRLHSA
Mvan_1301|M.vanbaalenii_PYR-1 --------------MSTPTMNQESQEAAKVLADP----TAYADDQRLHKA
MSMEG_1429|M.smegmatis_MC2_155 --------------MSTPTMD----DAAKALADP----TAYADDARLHEA
TH_4183|M.thermoresistible__bu --------------MSTPISSRLTDEAATVLADP----QAYTDEDSLNAA
MMAR_3999|M.marinum_M --------------MSAPTMD----EAA-LLADP----LAYTDETRLHAA
MUL_3863|M.ulcerans_Agy99 --------------MSAPTMD----EAA-LLADP----LAYTDETRLHAA
MAV_1609|M.avium_104 --------------MSTTTMD----EAAKLLADP----MAYTDEQRLHAA
MAB_3825|M.abscessus_ATCC_1997 --------------MATALSE--IEEAGRVLADP----AAYADELRLHAA
MLBr_02088|M.leprae_Br4923 MRTCVPTRTCVYAFIEYLSHNRPMGTNPPSLVEAQMLLLRLIDPGTRADP
: : * .
Mb2289|M.bovis_AF2122/97 FATLRREAPISFWPTIELPGFVAGNGHWALTKYDDVFYASRHPDIFSSYP
Rv2266|M.tuberculosis_H37Rv FATLRREAPISFWPTIELPGFVAGNGHWALTKYDDVFYASRHPDIFSSYP
Mflv_5053|M.gilvum_PYR-GCK LSYLRANDPVVWVDHPPYR------PFWAITKHADIMAIERANDLFLSAP
Mvan_1301|M.vanbaalenii_PYR-1 LAHLRANDPVAWVDHPPYR------PFWAITKHADIMAIERANDLFLSEP
MSMEG_1429|M.smegmatis_MC2_155 LARLRAENPVAWVDQAPYR------PFWAITKHADIMAIERANDLWLSAP
TH_4183|M.thermoresistible__bu LTHLRAHAPVSYVDVPNYR------PFWAITKHADIMDIERDNELWINAP
MMAR_3999|M.marinum_M LTRLRAQAPVALVDVPNYR------RFWAITRHADVMDIERDNTLFTNWP
MUL_3863|M.ulcerans_Agy99 LTRLRAQAPVALVDVPNYR------RFWAITRHADVMDIERDNTLFTNWP
MAV_1609|M.avium_104 LTHLRANAPVSWVEVPNYK------PFWAITKHADVMDIERENMLFTNWP
MAB_3825|M.abscessus_ATCC_1997 MTLLRREQPVTKVITDDYR------PFWAVTKHDDIMAVERDNALWINEP
MLBr_02088|M.leprae_Br4923 FPVYRALIDYGPMQLPGMP-------LTVFSSFSDCDEALRHPLSASDRL
:. * ..: . * * .
Mb2289|M.bovis_AF2122/97 NITINDQT--------PELAEYFGSMIVLDDPRHQRLRSIVSRAFTPKVV
Rv2266|M.tuberculosis_H37Rv NITINDQT--------PELAEYFGSMIVLDDPRHQRLRSIVSRAFTPKVV
Mflv_5053|M.gilvum_PYR-GCK RPLLVTAEADDLGKSQQEAGIGLRTLIHMDDPHHRKVRAIGADWFRPKAM
Mvan_1301|M.vanbaalenii_PYR-1 RPVLVTAEADDMARAQLEAGFGLRTLIHMDDPHHRKVRAIGADWFRPKAM
MSMEG_1429|M.smegmatis_MC2_155 RPLLATAEADDLGRSQQEMGIGLRTLIHMDDPHHRKVRAIGADWFRPKAM
TH_4183|M.thermoresistible__bu RPMLIPAATEDLAQANLAAGGGLRTLIHMDDPLHRDIRKIGADWFRPKAM
MMAR_3999|M.marinum_M RPVLATVAGDEL-----QAAAGVRTLIHLDDPQHRVARAIGADWFRPKAM
MUL_3863|M.ulcerans_Agy99 RPVLATVAGDEL-----QAAAGVRTLIHLDDPQHRVARAIGADWFRPKAM
MAV_1609|M.avium_104 RPVLTTAEGDEM-----QAAAGVRTLIHMDDPQHRVVRAIGSDWFRPKAM
MAB_3825|M.abscessus_ATCC_1997 RPLLMNLEQEAELDKQAAMGIELKTLVHIDDPKHRVLRAIGADWFRPKAM
MLBr_02088|M.leprae_Br4923 KATLAQQAIAAG---AEPRPFYASSFMFLDPPDHTRLRKLVSKAFAPKVV
. : ::: :* * * * : : * **.:
Mb2289|M.bovis_AF2122/97 ARIEAAVRDRAHRLVSSMIANNPDRQADLVSELAGPLPLQIICDMMGIPK
Rv2266|M.tuberculosis_H37Rv ARIEAAVRDRAHRLVSSMIANNPDRQADLVSELAGPLPLQIICDMMGIPK
Mflv_5053|M.gilvum_PYR-GCK RDLKVRVDELAKRYVDRMRDIGP--ECDFVTEIAVNFPLYVILSLLGLPE
Mvan_1301|M.vanbaalenii_PYR-1 RDLKIRVDELAKRYVDKMRDIGP--ECDFVTEIAVNFPLYVILSLLGLPE
MSMEG_1429|M.smegmatis_MC2_155 RELKVRVDELARIYVDKMREIGP--ECDFVTDIAVNFPLYVILSLLGLPE
TH_4183|M.thermoresistible__bu RALKTRVDELARIYVDKMVEVGP--ECNFVQEVAVNFPLYVILSLLGLPE
MMAR_3999|M.marinum_M RALKVRVDELAKNYVDAMMAAGP--ECDFVQQIAVNYPLYVIMSLLGLPE
MUL_3863|M.ulcerans_Agy99 RALKVRVDELAKNYVDAMMAAGP--ECDFVQQIAVNYPLYVIMSLLGLPE
MAV_1609|M.avium_104 RALKVRVDELAKIYVDKMLAAGP--ECDFVQEVAVNYPLYVIMSLLGLPE
MAB_3825|M.abscessus_ATCC_1997 RDMKLRTDELANRYVNKLLEAGG--SCDFAQDIAVHFPLYVIMSLLGIPE
MLBr_02088|M.leprae_Br4923 QALEGDIAALVDSLLDKGAAAGQ---FDVIADLAFPLAVAVICRLLGVPY
:: . :. . :. ::* .: :* ::*:*
Mb2289|M.bovis_AF2122/97 ADHQRIFHWTNVILGFGDPDLAT------DFDEFMQVSADIGAYATALAE
Rv2266|M.tuberculosis_H37Rv ADHQRIFHWTNVILGFGDPDLAT------DFDEFMQVSADIGAYATALAE
Mflv_5053|M.gilvum_PYR-GCK EDFGRMHMLTQEMFGGDD-EYKRG----TTPEEQLAVLTDFFNYFAALTA
Mvan_1301|M.vanbaalenii_PYR-1 EDFGRMHMLTQEMFGGDDDEYKRG----ATVEEQMAVLTDFFNYFGALTA
MSMEG_1429|M.smegmatis_MC2_155 EDFGRMHMLTQEMFGGDDDEYKRG----TTVEEQMAVLTDFFNYFSALTN
TH_4183|M.thermoresistible__bu SDFPRMLKLTQEMFGGDDDEFTRG----KSPQELHEVITDFFRYFTKLTA
MMAR_3999|M.marinum_M ADFPRMLKLTQELFGSDDSEFKRG----STSEDQLPALLDMFGYFNAVTT
MUL_3863|M.ulcerans_Agy99 ADFPRMLKLTQELFGSDDSEFKRG----STSEDQLPALLDMFGYFNAVTT
MAV_1609|M.avium_104 ADFPRMLKLTQELFGSDDSEFKRG----SSNEDQLPALLDMFGYFNGVTA
MAB_3825|M.abscessus_ATCC_1997 SDFDRMLKLTQELFGGDDSEFQRG----TTPEEQLMALLDFFGYFSGLTA
MLBr_02088|M.leprae_Br4923 EDAPEFGRVSALLVQSVDPFITITGEPPEATEERLRAGVWLRDYLEQLVK
* .: : :. * :: . : * :.
Mb2289|M.bovis_AF2122/97 DRRVNHHDDLTSSLVEAEVDGERLSSREIASFFILLVVAGNETTRNAITH
Rv2266|M.tuberculosis_H37Rv DRRVNHHDDLTSSLVEAEVDGERLSSREIASFFILLVVAGNETTRNAITH
Mflv_5053|M.gilvum_PYR-GCK SRREHPTEDLASAIANGLVDGEPMSDMDTLSYYVIVASAGHDTTKDAISG
Mvan_1301|M.vanbaalenii_PYR-1 SRRANPTDDLASAIANGLVDGELMSDVDTLSYYVIVASAGHDTTKDAISG
MSMEG_1429|M.smegmatis_MC2_155 SRRENPTDDLASAIANGRVDGELMSDMDTLSYYVIVASAGHDTTKDAISG
TH_4183|M.thermoresistible__bu DRRANPTEDLASAIANAKIDGEYLNDIDCLSYYVIVASAGHDTTSAAISG
MMAR_3999|M.marinum_M ARREHPTEDLASAIANARIDGEPLSDIDTVSYYLIVATAGHDTTSATISG
MUL_3863|M.ulcerans_Agy99 ARRAHPTADLASAIANARIDGEPLSDIDTVSYYLIVATAGHDTTSATISG
MAV_1609|M.avium_104 ARREHPTEDLASAIANARVDGEPLSDIDTVSYYLIVATAGHDTTSATISG
MAB_3825|M.abscessus_ATCC_1997 SRRQHPTDDLASTIANARVDGELLSDVDTASYYTIIATAGHDTTSATIAG
MLBr_02088|M.leprae_Br4923 CRRGTPGEDLISRLIELDESGDQLTEEEIIATCGLLLVAGHETTVNLIAN
** ** * : : .*: :.. : : :: **::** *:
Mb2289|M.bovis_AF2122/97 GVLALSRYPEQRDRWWSDFDGLAPTAVEEIVRWASPVVYMRRTLTQDIEL
Rv2266|M.tuberculosis_H37Rv GVLALSRYPEQRDRWWSDFDGLAPTAVEEIVRWASPVVYMRRTLTQDIEL
Mflv_5053|M.gilvum_PYR-GCK GLHALIENPGELARLKGDLS-LMPTAVEEMIRWSTPVKEFMRTAAEDTTV
Mvan_1301|M.vanbaalenii_PYR-1 GLHALVENPGELARLQGDLD-LMPTAVEEMIRWSTPVKEFMRTAAEDTTV
MSMEG_1429|M.smegmatis_MC2_155 GLHALIENPGELARLKADPG-LMGTAVEEMIRWSTPVKEFMRTAAEDTEV
TH_4183|M.thermoresistible__bu GLLALIENPDQLERLKAHPE-LMGTAVEEMIRWSTPVKEFMRTATRDTEV
MMAR_3999|M.marinum_M GLHALIEHPDQLARLQADPA-LIPLATEEMIRWVTPVKQFMRTASRDTVV
MUL_3863|M.ulcerans_Agy99 GLHALIEHPDQLARLQADPA-LMSLATEEMIRWVTPVKQFMRTASSDTVV
MAV_1609|M.avium_104 GLQALIENPDQLQRLRDNLD-LMPLATEEMIRWVTPVKEFMRTAAKDTVV
MAB_3825|M.abscessus_ATCC_1997 GLEALLEHPDQLARLRENPG-LMPLAVDEMIRWVTPVKEFMRTATADTEI
MLBr_02088|M.leprae_Br4923 AVLAMLRNPSQWKALSSNPQ-RAPLVVEETLRYDPAIHLIGRVAAKDMTI
.: *: . * : . ..:* :*: ..: : *. : * :
Mb2289|M.bovis_AF2122/97 RGTKMAAGDKVSLWYCSANRDESKFADPWTFDLARNPNPHLGFGGGGAHF
Rv2266|M.tuberculosis_H37Rv RGTKMAAGDKVSLWYCSANRDESKFADPWTFDLARNPNPHLGFGGGGAHF
Mflv_5053|M.gilvum_PYR-GCK RGVPIAKGESVYLAYVSANRDEEVFADPFRFDVGRDPNKHLAFGYG-VHF
Mvan_1301|M.vanbaalenii_PYR-1 RGVPIAKGESVYLAYVSANRDEDIFDDPFRFDVGRDPNKHLSFGYG-VHF
MSMEG_1429|M.smegmatis_MC2_155 RGVPIAKGESVYLAYVSGNRDEEVFTDPFRFDVGRDPNKHLAFGYG-VHF
TH_4183|M.thermoresistible__bu RGVPIKAGESVLLSYVSANRDEEVFDEPFRFDVGRDPNKHLSFGYG-VHF
MMAR_3999|M.marinum_M RGIPIAAGESVLLSYVSANRDEEVFDQPFRFDIGREPNKHVAFGYG-VHF
MUL_3863|M.ulcerans_Agy99 RGIPIAAGESVLLSYVSANRDEEVFDQPFRFDIGREPNKHVAFGYG-VHF
MAV_1609|M.avium_104 RGVPIAAGESVLLSYVSANRDEDVFDEPFRFDVGRDPNKHLAFGYG-VHF
MAB_3825|M.abscessus_ATCC_1997 RGVPITEGESVLLSYPSGNRDEDIFTDPFTFDIARDPNKHLAFGFG-VHF
MLBr_02088|M.leprae_Br4923 GQTTLTEGDTMVLLLAAANRDPAVYSRPDEFDPDRPSSRHLAFAVG-SHF
: *:.: * :.*** : * ** * .. *:.*. * **
Mb2289|M.bovis_AF2122/97 CLGANLARREIRVAFDELRRQMPDVVATEEPARLLSQFIHGIKTLPVTWS
Rv2266|M.tuberculosis_H37Rv CLGANLARREIRVAFDELRRQMPDVVATEEPARLLSQFIHGIKTLPVTWS
Mflv_5053|M.gilvum_PYR-GCK CLGAALARMEINSLFTELLPRLDSIELAGEAELSATTFVGGLKHLPVRYS
Mvan_1301|M.vanbaalenii_PYR-1 CLGAALARMEINSLFSELLPRLDSIELAGRPELSATTFVGGLKHLPVRYS
MSMEG_1429|M.smegmatis_MC2_155 CLGAALARMEMNSLFSELLPRLDSIELAGEPELSATTFVGGLKHLPIRYS
TH_4183|M.thermoresistible__bu CLGAALARMEINSFFTELVPRLESIELAGKPEFLATTFVGGLKHLPIRYS
MMAR_3999|M.marinum_M CMGAALARMEVGSFFTELLPRLKSIELSGDPQFTATTFVGGLKHLPVSYS
MUL_3863|M.ulcerans_Agy99 CMGAALARMEVASFFTELLPRLKSIELSGDPQFTATTFVGGLKRLPVSYS
MAV_1609|M.avium_104 CMGAALARMEVNSFFTELLPRLKSIELTGDPELVATTFVGGLKHLPVRYS
MAB_3825|M.abscessus_ATCC_1997 CLGAALARMEVNSFFSALLPRLESIEVDGNIERTSTIFVGGIKHLPIRYT
MLBr_02088|M.leprae_Br4923 CLGAALARLEATVTLSAISARFPQVQLAGELVYKPNVAMRGMSALPVQV-
*:** *** * : : :: .: . : *:. **:
Mb2289|M.bovis_AF2122/97 --
Rv2266|M.tuberculosis_H37Rv --
Mflv_5053|M.gilvum_PYR-GCK LR
Mvan_1301|M.vanbaalenii_PYR-1 LR
MSMEG_1429|M.smegmatis_MC2_155 IR
TH_4183|M.thermoresistible__bu VR
MMAR_3999|M.marinum_M LV
MUL_3863|M.ulcerans_Agy99 LM
MAV_1609|M.avium_104 LA
MAB_3825|M.abscessus_ATCC_1997 LR
MLBr_02088|M.leprae_Br4923 --