For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AGCRGPAIYTLLTMSSSCRYCRVHAGLHYRRVRAGQTGANVASARKLTAVAAISATLAQIVRLSISRSAF AGQASAANTELSQPSYVLLRVLIDEGPLPLSRLARLAHMDAGMATRRVRALVEAGLVTRHTDPNDGRVSV IEATPQGRRAAGALHEVRRDHLARALSGWSAAELHEFNRLLSKFLTDIKKTPIVDTATR*
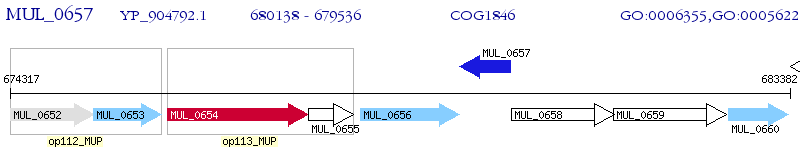
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_0657 | - | - | 100% (200) | transcriptional regulator |
| M. ulcerans Agy99 | MUL_1800 | - | 3e-07 | 28.03% (132) | transcriptional regulatory protein |
| M. ulcerans Agy99 | MUL_1238 | - | 9e-07 | 31.86% (113) | hypothetical protein MUL_1238 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0043c | - | 8e-10 | 32.50% (120) | MarR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_0856 | - | 5e-09 | 27.07% (133) | MarR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv0042c | - | 9e-10 | 32.50% (120) | MarR family transcriptional regulator |
| M. leprae Br4923 | MLBr_02696 | - | 1e-07 | 29.91% (107) | putative MarR-family regulatory protein |
| M. abscessus ATCC 19977 | MAB_1021 | - | 1e-06 | 28.57% (133) | MarR family transcriptional regulator |
| M. marinum M | MMAR_0904 | - | 1e-108 | 99.50% (200) | transcriptional regulator |
| M. avium 104 | MAV_0057 | - | 3e-09 | 33.94% (109) | transcriptional regulator, MarR family protein |
| M. smegmatis MC2 155 | MSMEG_4471 | - | 6e-09 | 30.20% (149) | MarR-family protein transcriptional regulator |
| M. thermoresistible (build 8) | TH_1373 | - | 2e-09 | 32.77% (119) | POSSIBLE TRANSCRIPTIONAL REGULATORY PROTEIN (POSSIBLY |
| M. vanbaalenii PYR-1 | Mvan_6049 | - | 6e-09 | 27.82% (133) | MarR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_0657|M.ulcerans_Agy99 ---------MAGCRGPAIYTLLTMSSSCR---------------------
MMAR_0904|M.marinum_M ---------MAGCRGPAIYTLLTMSSSCR---------------------
Mb0043c|M.bovis_AF2122/97 ----MSVVRSIGKKMQRISGPNALAVKG----------------------
Rv0042c|M.tuberculosis_H37Rv ----MSVVRSIGKKMQRISGPNALAVKG----------------------
MAV_0057|M.avium_104 --------------------------------------------------
MLBr_02696|M.leprae_Br4923 MFSYITTKSWNPGNQSRYSRPRRLSAYNGLYPKYFDVLDFTNDDPLHPAP
Mflv_0856|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_6049|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4471|M.smegmatis_MC2_155 --------------------------------------------------
TH_1373|M.thermoresistible__bu --------------------------------------------------
MAB_1021|M.abscessus_ATCC_1997 --------------------------------------------------
MUL_0657|M.ulcerans_Agy99 ------YCRVHAGLHYRRVRAGQTGANVASARKLTAVAAISATLAQIVRL
MMAR_0904|M.marinum_M ------YCRVHAGLHYRRVRAGRTGANVASARKLTAVAAISATLAQIVRL
Mb0043c|M.bovis_AF2122/97 ---RPTQVYGHT------HVRLDCRFMADSEFTAPEVTQLAEGLHRALSK
Rv0042c|M.tuberculosis_H37Rv ---RPTQVYGHT------HVRLDCRFMADSEFTAPEVTQLAEGLHRALSK
MAV_0057|M.avium_104 --------------------------MADSESNAAEVAELAEGLHRALSK
MLBr_02696|M.leprae_Br4923 PHLRPPLQLAHTRSLNARHIALNCRLMADSEFTAHEVTELADGLHRALSK
Mflv_0856|M.gilvum_PYR-GCK --------------------------MVT---TDAQVSELAGELQRVLSK
Mvan_6049|M.vanbaalenii_PYR-1 --------------------------MVP---TEARVSELASELQRVLSK
MSMEG_4471|M.smegmatis_MC2_155 ---------------------------------MTYETELGADLLAVVAR
TH_1373|M.thermoresistible__bu --------------------------------VDDSEARLASELSFAVVR
MAB_1021|M.abscessus_ATCC_1997 -------------------------MTVDDVDLDESLDALADSLLVASRL
:. *
MUL_0657|M.ulcerans_Agy99 SISRSAFAGQASAANTELSQPSYVLLRVLIDEGPLPLSRLARLAHMDAGM
MMAR_0904|M.marinum_M SISRSAFAGQASAANTELSQPSYVLLRVLIDEGPLPLSRLARLAHMDAGM
Mb0043c|M.bovis_AF2122/97 LISMLRRGDPNGAAAGDLTLAQLSILVTLLDQGPIRMTDLAAHERVRTPT
Rv0042c|M.tuberculosis_H37Rv LISMLRRGDPNGAAAGDLTLAQLSILVTLLDQGPIRMTDLAAHERVRTPT
MAV_0057|M.avium_104 LFAMLRRGDPSGAAAGDLTLAQLSILVTLLDQGPIRMTDLAAHERVRTPT
MLBr_02696|M.leprae_Br4923 LFSILRRRTPNGAVSGELTLAQLSILVTLLDQGPIRMTDLAAHERVRTPT
Mflv_0856|M.gilvum_PYR-GCK LMSVMRHTG-RTPTSGDLTLAQLSILLTLIDQGPMRMTELAAHEKVRTPT
Mvan_6049|M.vanbaalenii_PYR-1 VLSVMRHTG-RTATSGDLTLAQLSILLTLFDQGPMRMTELAAHEKVRTPT
MSMEG_4471|M.smegmatis_MC2_155 LNRLATQRT-----RLPLPYAQARLLSTIEDQGEARISDLALLDHCSQPT
TH_1373|M.thermoresistible__bu LARQLRARG----AESELSLAQLSALSTLVKEGAMTPGALAARERVRPPS
MAB_1021|M.abscessus_ATCC_1997 LLAIS--ARSIAQVDETITFPQFRTLVILFNEGPINLATLAQSLGVQPST
:. .. * : .:* **
MUL_0657|M.ulcerans_Agy99 ATRRVRALVEAGLVTRHTDPNDGRVSVIEATPQGRRAAGALHEVRRDHLA
MMAR_0904|M.marinum_M ATRRVRALVEAGLVTRHTDPNDGRVSVIEATPQGRRAAGALHEVRRDHLA
Mb0043c|M.bovis_AF2122/97 TTVAIRRLEKIGLVKRSRDPSDLRAVLVDITPQGRAVHGESLANRRAALA
Rv0042c|M.tuberculosis_H37Rv TTVAIRRLEKIGLVKRSRDPSDLRAVLVDITPQGRAVHGESLANRRAALA
MAV_0057|M.avium_104 TTVAIRRLEKVGLVKRSRDPSDLRAVLVDITPRGRAVHGESLANRRAALA
MLBr_02696|M.leprae_Br4923 TTVAIRRLEKIGLVKRSRDLSDLRAVLVDITPRGRTVHGESLANRRAALT
Mflv_0856|M.gilvum_PYR-GCK TTVAIRRLEKLGLVKRDRDPSDLRAVLVDITPEGLAQHQEALAIRQAHLA
Mvan_6049|M.vanbaalenii_PYR-1 TTVAIRRLEKLGLVKRDRDPSDLRAVLVDITPEGLAQHREALASRQAHLA
MSMEG_4471|M.smegmatis_MC2_155 MTTQVRRLEEAGLVSRTADPGDARAVLIRITEEGRKTLAQARADRAAAIN
TH_1373|M.thermoresistible__bu MTRVVSSLLELGLVARDAHPVDGRQVLIAVSAAGRRLVEAHRSTSQEWLM
MAB_1021|M.abscessus_ATCC_1997 TGRMVDRLVSAGLIDRRPHPRSRRELVAELTAHGEGVVREVMAQRRAEIA
: * . **: * . . * : : * :
MUL_0657|M.ulcerans_Agy99 RALSGWSAAELHEFNRLLSKFLTDIKKTPIVDTATR-------
MMAR_0904|M.marinum_M RALSGWSAAELHEFNRLLSKFLTDIKKTPIVDTATR-------
Mb0043c|M.bovis_AF2122/97 ALLSQLPRSDLETLRKALAPLERLASGEPASGPASNSPARKRA
Rv0042c|M.tuberculosis_H37Rv ALLSQLPRSDLETLRKALAPLERLASGEPASGPASNSPARKRA
MAV_0057|M.avium_104 AMLSQLPASDLDTLMKALAPLERLASGDPTSGPAGVPPARKEA
MLBr_02696|M.leprae_Br4923 TMLSQLPTSDLNILKNALAPLQRLASGEPVSSPAGDSPACKQA
Mflv_0856|M.gilvum_PYR-GCK SLLGKLSCDELDALTKALAPLERIAE-----------------
Mvan_6049|M.vanbaalenii_PYR-1 SLLSKLSDEELDALTKALAPLERIAE-----------------
MSMEG_4471|M.smegmatis_MC2_155 PRLERLSAEDRQTLVDAVEVIRKLLADVAEHPLPEEN------
TH_1373|M.thermoresistible__bu KRLAGLDAAQRETLTQAAELMAVLVDESA--------------
MAB_1021|M.abscessus_ATCC_1997 EIAARMPARERRGLVRALDAFTAAGGESASPVDV---------
: : :