For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TDKKIQVCVGIDVDSCAGWLGSYGGQDSPNDMQRGVFAGEVGVPRMLKLLERRGIVSTWFWPGHSIETFP TQAKMCVDAGHEIGAHGYSHENPRSLSQEQERAVMAKSVELVEQLTGARPKGYVAPWWEMSEHTATILKE YDFSYDHSQNYNDFVPFYARVGDEWTPIDTTKPAEHWMKPLKHGHEIDLVEFCGNWYVDDLPPMMFIKGH ANSHGFVNPRDIEQMWNDQFDWVYRELDYAVIPMTLHPDVSGRPQVLLMLERLLDRWAGFDGVEFVTMAD AAEQFRSRYPLDQPTRPECVGR*
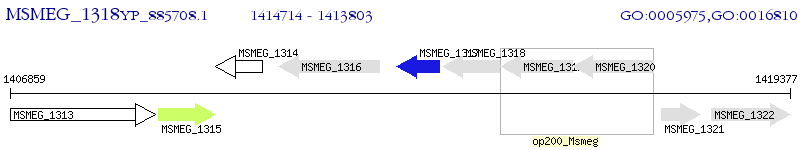
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1318 | - | - | 100% (303) | polysaccharide deacetylase family protein |
| M. smegmatis MC2 155 | MSMEG_1074 | - | 9e-30 | 32.28% (254) | polysaccharide deacetylase |
| M. smegmatis MC2 155 | MSMEG_4373 | - | 9e-28 | 28.57% (273) | polysaccharide deacetylase, putative |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1126 | - | 2e-05 | 27.85% (79) | glycosyl hydrolase |
| M. gilvum PYR-GCK | Mflv_0796 | - | 6e-29 | 31.23% (269) | polysaccharide deacetylase |
| M. tuberculosis H37Rv | Rv1096 | - | 2e-05 | 27.85% (79) | glycosyl hydrolase |
| M. leprae Br4923 | MLBr_01950 | - | 4e-07 | 26.23% (122) | carbohydrate degrading enzyme |
| M. abscessus ATCC 19977 | MAB_2935c | - | 4e-16 | 26.54% (260) | polysaccharide deacetylase family protein |
| M. marinum M | MMAR_1211 | - | 1e-17 | 27.13% (258) | chitinase |
| M. avium 104 | MAV_1218 | - | 2e-06 | 30.38% (79) | carbohydrate degrading enzyme |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_2681 | - | 3e-17 | 27.13% (258) | chitinase |
| M. vanbaalenii PYR-1 | Mvan_0047 | - | 1e-30 | 31.05% (277) | polysaccharide deacetylase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1126|M.bovis_AF2122/97 -----------------MPKRPDNQTWRYWRTVTGVVVAGAVLVVGGLSG
Rv1096|M.tuberculosis_H37Rv -----------------MPKRPDNQTWRYWRTVTGVVVAGAVLVVGGLSG
MAV_1218|M.avium_104 -----------------MAKRPDTLAWRYWRTVVGVLAAVAVLVIGGLTG
MLBr_01950|M.leprae_Br4923 -----------------MLKRPDTQAWRYSRTVFGVVAAGAVLVIGALTG
Mflv_0796|M.gilvum_PYR-GCK ------------------------MDAFRWPDGKTAAAAFTFDVDA--ES
Mvan_0047|M.vanbaalenii_PYR-1 -----------------------MMDRFRWPDGKTAAAAFTFDVDA--ES
MMAR_1211|M.marinum_M MTG----------YPRDMTGYGRTPPDPQWPGGALIAVQFVLNYEEGAES
MUL_2681|M.ulcerans_Agy99 MTG----------YPRDMTGYGRTPPDPQWRGGALIAVQFVLNYEEGAES
MAB_2935c|M.abscessus_ATCC_199 MAHNEAHDEAPPATPRDMVGYGRTPPHPRWPGNARIAVQFVLNYEEGAEN
MSMEG_1318|M.smegmatis_MC2_155 ------------------------------MTDKKIQVCVGIDVDSCAGW
. .
Mb1126|M.bovis_AF2122/97 RVTRAENLS-------------CSVIKCVALTFDDGPGPYTDRLLHILTD
Rv1096|M.tuberculosis_H37Rv RVTRAENLS-------------CSVIKCVALTFDDGPGPYTDRLLHILTD
MAV_1218|M.avium_104 HVTRADDLS-------------CSVVKCVALTFDDGPGPFDERLLQILKD
MLBr_01950|M.leprae_Br4923 HVTRADDMS-------------CAQVKCIALTFDDGPGPYTDRLVQILKD
Mflv_0796|M.gilvum_PYR-GCK AVLWGDES--------------VGARMSVMSHQAYGPLVGIPRILDLLEQ
Mvan_0047|M.vanbaalenii_PYR-1 AILWGNES--------------VGARMSVMSHQAYGPLVGIPRILDLLEQ
MMAR_1211|M.marinum_M SVLDGDSGSETFLSEIVPAQSFPNRHMSMESLYEYGSRAGVWRVLRVCEQ
MUL_2681|M.ulcerans_Agy99 SVLDGDSGSETFLSEIVPAQSFPNRHMSMESLYEYGSRAGVWRVLRVCEQ
MAB_2935c|M.abscessus_ATCC_199 NVLDGSAGSETFLSEMVPVEAFPNRHMSMESLYEYGSRAGLWRVLRVFER
MSMEG_1318|M.smegmatis_MC2_155 LGSYGGQDS-----------------PNDMQRGVFAGEVGVPRMLKLLER
. . *:: :
Mb1126|M.bovis_AF2122/97 NDAKATFFLIGNKVAANPAGARRIADAGMEIGSHTWEHPNMTTIPPEDIP
Rv1096|M.tuberculosis_H37Rv NDAKATFFLIGNKVAANPAGARRIADAGMEIGSHTWEHPNMTTIPPEDIP
MAV_1218|M.avium_104 NDAKATFFLIGNKVAANPAGAKRIADAGMEIGNHTWEHPNMTTIPTEDVA
MLBr_01950|M.leprae_Br4923 NDAKATFFLIGNKVATNPAGAKRIVDAGMEIGSHTWEHPNMTMLSTEDIA
Mflv_0796|M.gilvum_PYR-GCK HQITSTFFVPGHTADRYPEAVRSIVGAGHEIAHHGYLHEQPTALTLEEEI
Mvan_0047|M.vanbaalenii_PYR-1 HQVPSTFFVPGHTADRYPEAVRSIVAAGHEIAHHGYLHEQPTALTLEQEI
MMAR_1211|M.marinum_M SGVPLTIFAVAQAMQRNPEVAKVFRELGHEIACHGLRWKSYQEIDRDTER
MUL_2681|M.ulcerans_Agy99 SGVPLTIFAVAQAMQRNPEVAKGFRELGHEIACHGLRWKSYQEIDRDTER
MAB_2935c|M.abscessus_ATCC_199 RGLPLTIFAVATALQRNPEAVSAFGELGHEIACHGLRWQSYQLVPPDIER
MSMEG_1318|M.smegmatis_MC2_155 RGIVSTWFWPGHSIETFPTQAKMCVDAGHEIGAHGYSHENPRSLSQEQER
* * . * . * **. * . : :
Mb1126|M.bovis_AF2122/97 GQFSRANDVIAAATGRTPTLYRP---AGGLSNDAVRQAAAKVGQAEI-LW
Rv1096|M.tuberculosis_H37Rv GQFSRANDVIAAATGRTPTLYRP---AGGLSNDAVRQAAAKVGQAEI-LW
MAV_1218|M.avium_104 AQFTRANDAIHAATGRTPNLYRP---AGGLSNPVVRQTAGQLGLAEI-LW
MLBr_01950|M.leprae_Br4923 DQFSRANHAITVATGRTPTLWRP---AGGLSNEAVRQVAGRYGQAEI-LW
Mflv_0796|M.gilvum_PYR-GCK DALDRGLAALADVAGVRPAGYRAPMWDLSWRTPALLAERGFLYDSSL-MD
Mvan_0047|M.vanbaalenii_PYR-1 DALDRGLAALADVAGVRPVGYRAPMWDLSWRTPALLAERDFLYDSSL-MD
MMAR_1211|M.marinum_M AHLSEAVRIHHELTGSAPRGWYG-GRDSPHTRELVVEHGGFLYDSDS-YA
MUL_2681|M.ulcerans_Agy99 AHLSEAVRIHHELTGSAPRGWYG-GRDSPHTRELVVEHGGFLYDSDS-YA
MAB_2935c|M.abscessus_ATCC_199 EHMAQAVQILRDLTGQAPLGWYT-GRDSPQTRQLVVEHGGFSYDSDS-YA
MSMEG_1318|M.smegmatis_MC2_155 AVMAKSVELVEQLTGARPKGYVAPWWEMSEHTATILKEYDFSYDHSQNYN
: .. :* * : : .
Mb1126|M.bovis_AF2122/97 DVIPFDWIN------------------------DSNTAATRHMLMTQIKP
Rv1096|M.tuberculosis_H37Rv DVIPFDWIN------------------------DSNTAATRHMLMTQIKP
MAV_1218|M.avium_104 DVIPFDWAN------------------------DSNTAATRYMLMTYIKP
MLBr_01950|M.leprae_Br4923 DVIPFDWTN------------------------DSDTVATRCMLMMQIKP
Mflv_0796|M.gilvum_PYR-GCK ADHPYELAVTPG--------------------AAQSLVEIPIQWALDDWE
Mvan_0047|M.vanbaalenii_PYR-1 ADHPYELAITPG--------------------AQESLVEIPIQWALDDWE
MMAR_1211|M.marinum_M DDLPYWVRV-----------------------GASDHLVVPYTLDTNDMR
MUL_2681|M.ulcerans_Agy99 DDLPYWVRV-----------------------GASDHLVVPYTLDTNDMR
MAB_2935c|M.abscessus_ATCC_199 DDLPYWVRVPAPG-------------------GVADHLVVPYTLDTNDMR
MSMEG_1318|M.smegmatis_MC2_155 DFVPFYARVGDEWTPIDTTKPAEHWMKPLKHGHEIDLVEFCGNWYVDDLP
*: .
Mb1126|M.bovis_AF2122/97 GSVVLFHDTYSSTVDVVYQFIPVLKANGYRLVTVS--ELLGPRAPGSSYG
Rv1096|M.tuberculosis_H37Rv GSVVLFHDTYSSTVDVVYQFIPVLKANGYRLVTVS--ELLGPRAPGSSYG
MAV_1218|M.avium_104 GSVVLFHNTYSSTVDLVYQFIPVLKANGYRLVTVS--ELLGPRAPGSSYG
MLBr_01950|M.leprae_Br4923 GSVVLFHDTYSSTVDLVYQFIPVLKANGYRLVTVT--ELLGPRAPGSSYG
Mflv_0796|M.gilvum_PYR-GCK QYCFLPDISGSGLIESPRKARELWQLEFDALRRVG--GCWVLTNHPFLTG
Mvan_0047|M.vanbaalenii_PYR-1 QYCFLPDISGSGLIETPRKARELWQSEFDALRKVG--GCWVLTNHPFLTG
MMAR_1211|M.marinum_M FASPAGFSNGDEFFAHLRDAFDVLYAEGRAGSPK----MLSVGLHCRLVG
MUL_2681|M.ulcerans_Agy99 FASPAGFSNGDEFFAHLRDAFDVLYAEGRAGSPK----MLSVGLHCRLVG
MAB_2935c|M.abscessus_ATCC_199 FSSPAGFPSGEQFFTHLRDAFDCLYAEGEAGSPK----MLSVGLHCRLVG
MSMEG_1318|M.smegmatis_MC2_155 PMMFIKGHANSHGFVNPRDIEQMWNDQFDWVYRELDYAVIPMTLHPDVSG
. . . : *
Mb1126|M.bovis_AF2122/97 SRENGPPVNELRDIPASEIPPLPNTSSPKPMSNFPITDIAGQNSGGPNNG
Rv1096|M.tuberculosis_H37Rv SRENGPPVNELRDIPASEIPPLPNTSSPKPMPNFPITDIAGQNSGGPNNG
MAV_1218|M.avium_104 SRENGPPVNGLRDIPASDIPKLPDTPSPKPMPNFPITDIPGQNSGGPNNG
MLBr_01950|M.leprae_Br4923 RRENGPPVNQLHDIPANLIPALPNTSSPKPMPNFPITDISGQNSGGSNDG
Mflv_0796|M.gilvum_PYR-GCK RPSRAAELGELMRYVLDHDDVWVVSQA-----------------------
Mvan_0047|M.vanbaalenii_PYR-1 RPSRAAELGELMRYVLDHDDVWVSSLGAVAEHVRSLNLSPRSITPPDVPR
MMAR_1211|M.marinum_M RPARTAALQRFLDHVRSHDRVWLARRVEIAEHWREVHPAPTG--------
MUL_2681|M.ulcerans_Agy99 RPARTAALQRFLDHVRSHDRVWLARRVEIAEHWREVHPAPTG--------
MAB_2935c|M.abscessus_ATCC_199 RPARTVALERFVDHVQAHDGVWITKRIDIAEHWRRVHPAGTEPNESSSPA
MSMEG_1318|M.smegmatis_MC2_155 RPQVLLMLERLLDRWAGFDGVEFVTMADAAEQFRSRYPLDQPTRPECVGR
: :
Mb1126|M.bovis_AF2122/97 A
Rv1096|M.tuberculosis_H37Rv A
MAV_1218|M.avium_104 A
MLBr_01950|M.leprae_Br4923 A
Mflv_0796|M.gilvum_PYR-GCK -
Mvan_0047|M.vanbaalenii_PYR-1 -
MMAR_1211|M.marinum_M -
MUL_2681|M.ulcerans_Agy99 -
MAB_2935c|M.abscessus_ATCC_199 -
MSMEG_1318|M.smegmatis_MC2_155 -