For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
YTTRRNLLFGTAATIAAMTTGACSSSAAQQPQAQPSAPPTPSFQPNQRYPDPAIEILDPSFAKYRLFSSS LEKVATGLRWAEGPVYFPDGGYLLCSDTPNNRLMKFDEKDGSFTVFRHPSNYSNGNTRDRQGRLVTCEHS ETRRITRTEENGDITVLADSFEGKRLNAPNDIVVKSDDTIWFTDPLFGINGEWEGSRAEPEQATTNVYRL TPDGELTAVITDLVNPNGIGFSPDEKKLYVIEWRGTPNRSLWSFDVNDDGTVANKTKLIDAADQGALDGF TVDRDGNLWCGWGSNGLVADQPTDVGGRKVYELRGKPEDLDGVMVFNPQGKPIGFIRLPERCANLTFGGP ENNRLYMAASHSLYALYVEAFGAT*
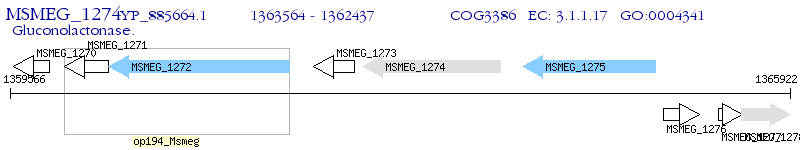
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1274 | - | - | 100% (375) | gluconolactonase |
| M. smegmatis MC2 155 | MSMEG_6378 | - | 1e-09 | 25.85% (294) | senescence marker protein-30 |
| M. smegmatis MC2 155 | MSMEG_4005 | - | 6e-08 | 30.09% (113) | calcium-binding protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4863 | - | 8e-08 | 24.15% (207) | strictosidine synthase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3680 | - | 2e-05 | 23.75% (160) | strictosidine synthase family protein |
| M. marinum M | MMAR_0391 | - | 2e-05 | 34.94% (83) | hypothetical protein MMAR_0391 |
| M. avium 104 | MAV_5132 | - | 1e-08 | 37.35% (83) | senescence marker protein-30 (SMP-30) |
| M. thermoresistible (build 8) | TH_3744 | - | 1e-11 | 25.77% (326) | PUTATIVE - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_1568 | - | 2e-07 | 24.80% (246) | strictosidine synthase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0391|M.marinum_M --------------------------------------------------
MAV_5132|M.avium_104 --------------------------------------------------
Mflv_4863|M.gilvum_PYR-GCK --------------------------------------------MPRFKR
Mvan_1568|M.vanbaalenii_PYR-1 -----------------------------------------------MKP
MAB_3680|M.abscessus_ATCC_1997 ----------------------------------------------MAKP
TH_3744|M.thermoresistible__bu -----------------------------------------------LAF
MSMEG_1274|M.smegmatis_MC2_155 MYTTRRNLLFGTAATIAAMTTGACSSSAAQQPQAQPSAPPTPSFQPNQRY
MMAR_0391|M.marinum_M ------MHDTFAVICNGTRREVSPLARGFCFGEGPR----WFEGLLWFSD
MAV_5132|M.avium_104 ------------MIP-------EPLVNGFCFGEGPR----WFEGLLWFSD
Mflv_4863|M.gilvum_PYR-GCK PIDPVRWQAPPVDPLPEFG--SAALTLTPIPGGEPEDVVVDARGYLWTGA
Mvan_1568|M.vanbaalenii_PYR-1 SIDPVRWHAPPVQDLPEFG--AAELTLVPVPGGEPEDVVVDAEGNLWTGA
MAB_3680|M.abscessus_ATCC_1997 SIDPVRWQPPGVRALSLEPQPLPRLSIVALPGHGPEDVVADAAGNIWAGV
TH_3744|M.thermoresistible__bu PESPIAEADGSVLVSEIAG---GCITRVRPDGTTEKVADTGGGPNGIARL
MSMEG_1274|M.smegmatis_MC2_155 PDPAIEILDPSFAKYRLFSSSLEKVATGLRWAEGPVYFP-DGGYLLCSDT
: .
MMAR_0391|M.marinum_M MLGEAVHTSDM---HGSLTTLPLPGHSPCGLGFRPDGTLLIASAHD-RLV
MAV_5132|M.avium_104 MLGEAVHTTTM---GGALTTLPLPGHCPSGLGFRPDGSLLIASTRD-RRV
Mflv_4863|M.gilvum_PYR-GCK LDGSIVRLRPD---GTAPEVVANTGGRPLGLAFARDGRLLVCDSPRGLLA
Mvan_1568|M.vanbaalenii_PYR-1 LDGGIVRLRPD---GSSPELIANTGGRPLGLTFARDGRLLVCDSPRGLLA
MAB_3680|M.abscessus_ATCC_1997 ADGRIFRISPDDTEGAAVTHVATTEHPPLGLHIARDGRVLICSRDK-LLA
TH_3744|M.thermoresistible__bu PDGRLVVCQNG---GSTFGVGSWPYDLPGATKLFRPIGPATAPLTPQLQV
MSMEG_1274|M.smegmatis_MC2_155 PNNRLMKFDEK---DGSFTVFRHPSNYSNGNTRDRQGRLVTCEHSETRRI
. . : . . . .
MMAR_0391|M.marinum_M LRYDG--DTVVTVADLRDLAPADLGDMVIDRAG--------------RAY
MAV_5132|M.avium_104 LRYDG--ETVVTVAELADIAPADLGDMVVDRAG--------------RAY
Mflv_4863|M.gilvum_PYR-GCK LDVDT--GAIETLVTSIDGRPLLFCSNVTETSDGTVYFTESTSAFTIDQY
Mvan_1568|M.vanbaalenii_PYR-1 VDTTT--GTVETLVHSVGGRPLIFCSNVTETADGTVYFTESTTAFTVGNY
MAB_3680|M.abscessus_ATCC_1997 LDPAS--GKIEPVVTKVDGPPLIFCSNVTESMDGTIYFSESTARFPFEQF
TH_3744|M.thermoresistible__bu IDADGRIETLTTEFIARNGQRLPLVRPSDVCVD-------SSGGYYVTDG
MSMEG_1274|M.smegmatis_MC2_155 TRTEEN-GDITVLADSFEGKRLNAPNDIVVKSDDTIWFTDPLFGINGEWE
: .
MMAR_0391|M.marinum_M IGCQSFSG--GVIIRLDTDNSAQVVAEDLDFPNGMVITPDHDTLIVAESV
MAV_5132|M.avium_104 IGCQAFTR--GAIIRLDPDDRAKVVADDLDFPNGMAITPDGATLIVAEST
Mflv_4863|M.gilvum_PYR-GCK LGAILEARGRGALHRLDPDGRVTTVVDGLYFANGVTPTADGSALVFAETQ
Mvan_1568|M.vanbaalenii_PYR-1 LGAILEARGRGALHRLEPHGRLTTVVDGLYFANGVTPTADGSALVFAETQ
MAB_3680|M.abscessus_ATCC_1997 MAAILEGRPTGRVFRRDPDGTVTTIATGLAFTNGVTITADGSALIIAETV
TH_3744|M.thermoresistible__bu GTIRGRSREMTGLLYAGLDGVLREIVYPLEMPNGVALSPDESKLYVAETR
MSMEG_1274|M.smegmatis_MC2_155 GSRAEPEQATTNVYRLTPDGELTAVITDLVNPNGIGFSPDEKKLYVIEWR
: .. : * .**: :.* * . *
MMAR_0391|M.marinum_M G---RRLSAFTVSADGALNDRRVFAAGLDG-PPDGIALDAEGAVWAAMTL
MAV_5132|M.avium_104 G---RRLSAFAIDDDGALSGRRVFADGLDG-PPDGIALDADGGVWAAMTL
Mflv_4863|M.gilvum_PYR-GCK G---RRLSKYWLTGPNAGS-VTPLAVNLPA-MPDNLSTGAEGRIWCAMVT
Mvan_1568|M.vanbaalenii_PYR-1 G---RRLSKYWLTGPDAGN-VTPLAVNLPA-MPDNLSTGADGRIWCAMVT
MAB_3680|M.abscessus_ATCC_1997 G---RRVSRYALTGPAAGT-LTPIVEEIPG-MPDNISTGADGRIWITLAS
TH_3744|M.thermoresistible__bu T---RRIWEFGLDEPGVIG----SARGLAT-VPSGGPLNVGGADGLCVDS
MSMEG_1274|M.smegmatis_MC2_155 GTPNRSLWSFDVNDDGTVANKTKLIDAADQGALDGFTVDRDGNLWCGWGS
* : : : . .. . . *
MMAR_0391|M.marinum_M AH--QFERIVAGG--------VVTDRIDMG-DRVAIACALGGPGRRTLFL
MAV_5132|M.avium_104 AH--QFERIVAGG--------AVTDRIDIG-DRVAIACALGGPQRRTLFL
Mflv_4863|M.gilvum_PYR-GCK PANPVADRLAAGPPVLRKLVWRLPARLQPKPEAVAWAVAFDPDSGDAVAG
Mvan_1568|M.vanbaalenii_PYR-1 PANPLADRLAAGPPLLRKLVWRLPSRVQPKPEAVVWVVAFDPDTGAAVAG
MAB_3680|M.abscessus_ATCC_1997 PRNALAEWLLPRSPAIRKVLWRLPDALLPGTDTDPWVIAVNPDTGDVLAN
TH_3744|M.thermoresistible__bu AGRILVATLGTGG-------------VTVFSAAGELLGAIEAD--DPMTT
MSMEG_1274|M.smegmatis_MC2_155 NGLVADQPTDVGG--------RKVYELRGKPEDLDGVMVFNPQ-GKPIGF
: .. :
MMAR_0391|M.marinum_M LSSTDAYPKRLIGTRLS--QLDAVTVATPGAGLP------------
MAV_5132|M.avium_104 LSSTDAYPQRLVGTRLS--RLDAVTVTTPGAGLP------------
Mflv_4863|M.gilvum_PYR-GCK FRTTHPEFRMATGLVESGGRLWLGSIGGPYLASVDLAATAP-----
Mvan_1568|M.vanbaalenii_PYR-1 MRTTHPDFSMVTGVAEAGGRLWMGSIGSPNLGWITL----------
MAB_3680|M.abscessus_ATCC_1997 ITGKSRDLRTVTGVVESGGRLWMGCIGSSAVGHISLADIASSSSGP
TH_3744|M.thermoresistible__bu NMTLSPDETRLYVTLASSGRILVVEDWMTAAGIR------------
MSMEG_1274|M.smegmatis_MC2_155 IRLPERCANLTFGGPENNRLYMAASHSLYALYVEAFGAT-------