For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAKPSIDPVRWQPPGVRALSLEPQPLPRLSIVALPGHGPEDVVADAAGNIWAGVADGRIFRISPDDTEGA AVTHVATTEHPPLGLHIARDGRVLICSRDKLLALDPASGKIEPVVTKVDGPPLIFCSNVTESMDGTIYFS ESTARFPFEQFMAAILEGRPTGRVFRRDPDGTVTTIATGLAFTNGVTITADGSALIIAETVGRRVSRYAL TGPAAGTLTPIVEEIPGMPDNISTGADGRIWITLASPRNALAEWLLPRSPAIRKVLWRLPDALLPGTDTD PWVIAVNPDTGDVLANITGKSRDLRTVTGVVESGGRLWMGCIGSSAVGHISLADIASSSSGP
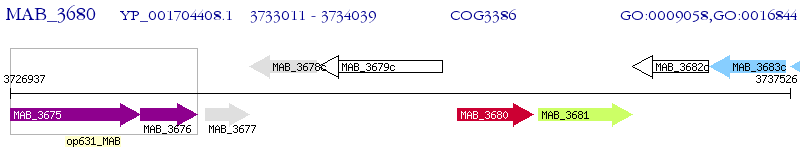
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3680 | - | - | 100% (342) | strictosidine synthase family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4863 | - | 4e-93 | 50.88% (340) | strictosidine synthase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_0391 | - | 7e-10 | 26.86% (175) | hypothetical protein MMAR_0391 |
| M. avium 104 | MAV_5132 | - | 5e-13 | 28.49% (179) | senescence marker protein-30 (SMP-30) |
| M. smegmatis MC2 155 | MSMEG_2561 | - | 1e-10 | 30.11% (186) | gluconolactonase |
| M. thermoresistible (build 8) | TH_0278 | - | 3e-12 | 29.38% (177) | gluconolactonase |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_1568 | - | 9e-92 | 51.36% (331) | strictosidine synthase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0391|M.marinum_M ------------MHDTFAVICNGTRREVSPLARGFCFGEGPR----WFEG
MAV_5132|M.avium_104 ------------------MIP-------EPLVNGFCFGEGPR----WFEG
TH_0278|M.thermoresistible__bu ------------------VLN------PTPLAKGFCFGEGPR----WFEG
MSMEG_2561|M.smegmatis_MC2_155 ------------------MTTEAGPERTTELATGFCFGEGPR----WFEG
Mflv_4863|M.gilvum_PYR-GCK MPRFKRPIDPVRWQAPPVDPLPEFG--SAALTLTPIPGGEPEDVVVDARG
Mvan_1568|M.vanbaalenii_PYR-1 ---MKPSIDPVRWHAPPVQDLPEFG--AAELTLVPVPGGEPEDVVVDAEG
MAB_3680|M.abscessus_ATCC_1997 --MAKPSIDPVRWQPPGVRALSLEPQPLPRLSIVALPGHGPEDVVADAAG
* * *. *
MMAR_0391|M.marinum_M LLWFSDMLGEAVHTSDMH---GSLTTLPLPGHSPCGLGFRPDGTLLIASA
MAV_5132|M.avium_104 LLWFSDMLGEAVHTTTMG---GALTTLPLPGHCPSGLGFRPDGSLLIAST
TH_0278|M.thermoresistible__bu LLWFSDMLGEAVHTVDLN---GSMTTLPLPGHAPSGLGFRPDGSLLIAST
MSMEG_2561|M.smegmatis_MC2_155 LLWFSDMLGEAVHTVTLG---GSMTTLPLPGHAPSGLGFRPDGTLYIVST
Mflv_4863|M.gilvum_PYR-GCK YLWTGALDGSIVRLRPD---GTAPEVVANTGGRPLGLAFARDGRLLVCDS
Mvan_1568|M.vanbaalenii_PYR-1 NLWTGALDGGIVRLRPD---GSSPELIANTGGRPLGLTFARDGRLLVCDS
MAB_3680|M.abscessus_ATCC_1997 NIWAGVADGRIFRISPDDTEGAAVTHVATTEHPPLGLHIARDGRVLICSR
:* . * .: : :. . * ** : ** : : .
MMAR_0391|M.marinum_M HD-RLVLRYDGDTVVTVADLRDLAPADLGDMVID-RAGRAYIGC------
MAV_5132|M.avium_104 RD-RRVLRYDGETVVTVAELADIAPADLGDMVVD-RAGRAYIGC------
TH_0278|M.thermoresistible__bu EA-RCVLRYDGETVTTIADLSAFAPDNLGDMVVD-DAGRAYVGS------
MSMEG_2561|M.smegmatis_MC2_155 EK-RQVLCYDGETVELLADLHQVAPAPLGDMVLD-ERGRAYVGS------
Mflv_4863|M.gilvum_PYR-GCK PRGLLALDVDTGAIETLVTSIDGRPLLFCSNVTETSDGTVYFTESTSAFT
Mvan_1568|M.vanbaalenii_PYR-1 PRGLLAVDTTTGTVETLVHSVGGRPLIFCSNVTETADGTVYFTESTTAFT
MAB_3680|M.abscessus_ATCC_1997 DK-LLALDPASGKIEPVVTKVDGPPLIFCSNVTESMDGTIYFSESTARFP
.: : :. * : . * : * *.
MMAR_0391|M.marinum_M ---------QSFSGGVIIRLDTDNS---AQVVAEDLDFPNGMVITPDHDT
MAV_5132|M.avium_104 ---------QAFTRGAIIRLDPDDR---AKVVADDLDFPNGMAITPDGAT
TH_0278|M.thermoresistible__bu ---------QAYSGGVIVRVDPDDPDDPVTVVAEDLDFPNGMVITEDRST
MSMEG_2561|M.smegmatis_MC2_155 ---------QARGGGVIVRIDPDDPVEPIRVVAEGLDFPNGMAITPDGET
Mflv_4863|M.gilvum_PYR-GCK IDQYLGAILEARGRGALHRLDPDGR---VTTVVDGLYFANGVTPTADGSA
Mvan_1568|M.vanbaalenii_PYR-1 VGNYLGAILEARGRGALHRLEPHGR---LTTVVDGLYFANGVTPTADGSA
MAB_3680|M.abscessus_ATCC_1997 FEQFMAAILEGRPTGRVFRRDPDGT---VTTIATGLAFTNGVTITADGSA
:. * : * :... .:. .* *.**:. * * :
MMAR_0391|M.marinum_M LIVAESVGRRLSAFTVSADGALNDRRVFAAGLDGPPDGIALDAEGAVWAA
MAV_5132|M.avium_104 LIVAESTGRRLSAFAIDDDGALSGRRVFADGLDGPPDGIALDADGGVWAA
TH_0278|M.thermoresistible__bu LIVAESIGRRLTSYRIAADGTLTDREVFADGLDGPPDGIALDAAGAVWTS
MSMEG_2561|M.smegmatis_MC2_155 LIVAESTARRLTAYTVDEDGDLSDRRIFAEGLDGPPDGIAIDLEGGVWTA
Mflv_4863|M.gilvum_PYR-GCK LVFAETQGRRLSKYWLTG-PNAGSVTPLAVNLPAMPDNLSTGAEGRIWCA
Mvan_1568|M.vanbaalenii_PYR-1 LVFAETQGRRLSKYWLTG-PDAGNVTPLAVNLPAMPDNLSTGADGRIWCA
MAB_3680|M.abscessus_ATCC_1997 LIIAETVGRRVSRYALTG-PAAGTLTPIVEEIPGMPDNISTGADGRIWIT
*:.**: .**:: : : :. : . **.:: . * :* :
MMAR_0391|M.marinum_M MTLAHQ--FERIVAGG--------VVTDRIDMG-DRVAIACALGGPGRRT
MAV_5132|M.avium_104 MTLAHQ--FERIVAGG--------AVTDRIDIG-DRVAIACALGGPQRRT
TH_0278|M.thermoresistible__bu MTLAHR--FDRILPGG--------EVTDRIDIG-GRAAIACALGGPGRNL
MSMEG_2561|M.smegmatis_MC2_155 LTLAHE--FQRIVDSGPGT----PTVTDRIDAG-GRTAIACALGGAEGRT
Mflv_4863|M.gilvum_PYR-GCK MVTPANPVADRLAAGPPVLRKLVWRLPARLQPKPEAVAWAVAFDPDSGDA
Mvan_1568|M.vanbaalenii_PYR-1 MVTPANPLADRLAAGPPLLRKLVWRLPSRVQPKPEAVVWVVAFDPDTGAA
MAB_3680|M.abscessus_ATCC_1997 LASPRNALAEWLLPRSPAIRKVLWRLPDALLPGTDTDPWVIAVNPDTGDV
:. . . : : :. : . *..
MMAR_0391|M.marinum_M LFLLSSTDAYPKRLIGTRLS--QLDAVTVATPGAG-LP-----------
MAV_5132|M.avium_104 LFLLSSTDAYPQRLVGTRLS--RLDAVTVTTPGAG-LP-----------
TH_0278|M.thermoresistible__bu LFLLSTTDAYPKRLIGTQSA--RLDVVKVDVAGAG-LP-----------
MSMEG_2561|M.smegmatis_MC2_155 LFLVTTTDAYPQRLVGTKLS--RIEALDVAVPAPGDFAVGDHHHH----
Mflv_4863|M.gilvum_PYR-GCK VAGFRTTHPEFRMATGLVESGGRLWLGSIGGPYLASVDLAATAP-----
Mvan_1568|M.vanbaalenii_PYR-1 VAGMRTTHPDFSMVTGVAEAGGRLWMGSIGSPNLGWITL----------
MAB_3680|M.abscessus_ATCC_1997 LANITGKSRDLRTVTGVVESGGRLWMGCIGSSAVGHISLADIASSSSGP
: . . * : :: : . . .