For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MGTAINRDYSITVVADLKTTLGEGPLWDTEQQLLYWLDSADGRIFRATADGTQIRAWDVREKIGSMTLYR DGTAALTALENGLYRVDFETGEQTEVLELERDLPSNRLNDGKVDKRGRFVFGSMDTLEESRSGKLYSYGP DGTLQILDEDITVSNGPCWSPDGRTLYFCDTWTGEIWCYDYDLDTGKVSNRRTFARVDTSGGGAADGATV DAEGYLWQALVYGGKIIRYAPDGTVDHVIDFPVLKPTSVMFGGPDLDVLYVTSMARPPLPRFPEDGQLRG SLFAIHGLGVRGVPEPRFGA
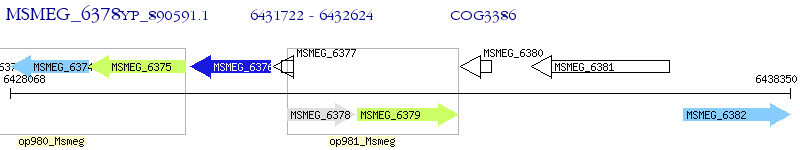
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6378 | - | - | 100% (300) | senescence marker protein-30 |
| M. smegmatis MC2 155 | MSMEG_4005 | - | 2e-25 | 36.81% (182) | calcium-binding protein |
| M. smegmatis MC2 155 | MSMEG_2561 | - | 5e-11 | 26.10% (249) | gluconolactonase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4759 | - | 8e-29 | 32.58% (264) | SMP-30/gluconolaconase/LRE domain-containing protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_0391 | - | 5e-11 | 25.93% (243) | hypothetical protein MMAR_0391 |
| M. avium 104 | MAV_5132 | - | 8e-13 | 27.05% (244) | senescence marker protein-30 (SMP-30) |
| M. thermoresistible (build 8) | TH_0278 | - | 5e-10 | 25.51% (247) | gluconolactonase |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_2237 | - | 4e-11 | 24.90% (261) | SMP-30/gluconolaconase/LRE domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0391|M.marinum_M MHDTFAVICNGTRREVSPLARGFCFGEGPRW--FEGLLWFSDMLGEAVHT
MAV_5132|M.avium_104 ------MIP-------EPLVNGFCFGEGPRW--FEGLLWFSDMLGEAVHT
TH_0278|M.thermoresistible__bu ------VLN------PTPLAKGFCFGEGPRW--FEGLLWFSDMLGEAVHT
Mvan_2237|M.vanbaalenii_PYR-1 -------MT------LTSLANGFCFGEGPRW--FEGLVWFSDMLGEAVHT
Mflv_4759|M.gilvum_PYR-GCK -------MTR---CAERLTPRAAHHGEGPFWDRRSGRLLFMDVLAGVVGA
MSMEG_6378|M.smegmatis_MC2_155 ----MGTAINRDYSITVVADLKTTLGEGPLWDTEQQLLYWLDSADGRIFR
**** * . : : * :
MMAR_0391|M.marinum_M SDMHG-SLTTLPLPGHSPCGLGFRPDGTLLIASAHDRLVLRYDGDTVVTV
MAV_5132|M.avium_104 TTMGG-ALTTLPLPGHCPSGLGFRPDGSLLIASTRDRRVLRYDGETVVTV
TH_0278|M.thermoresistible__bu VDLNG-SMTTLPLPGHAPSGLGFRPDGSLLIASTEARCVLRYDGETVTTI
Mvan_2237|M.vanbaalenii_PYR-1 VTLAG-EMSTLQLAGHAPSGLGFRPDGSLLIVSTERRQILGYDGDSVTTV
Mflv_4759|M.gilvum_PYR-GCK IESTG-KVLRYDVPADVATVIRRRASGGFVIATGHDLMCADDDLANFTHL
MSMEG_6378|M.smegmatis_MC2_155 ATADGTQIRAWDVREKIGSMTLYRDGTAALTALENGLYRVDFETGEQTEV
* : : . * . : . . : . :
MMAR_0391|M.marinum_M ADLR-DLAPADLGDMVIDRAGRAYIGCQSF----SGGVIIRLDTDNS---
MAV_5132|M.avium_104 AELA-DIAPADLGDMVVDRAGRAYIGCQAF----TRGAIIRLDPDDR---
TH_0278|M.thermoresistible__bu ADLS-AFAPDNLGDMVVDDAGRAYVGSQAY----SGGVIVRVDPDDPDDP
Mvan_2237|M.vanbaalenii_PYR-1 ADLS-AMVPAALGDMVIDRHGGAYVGSQAR----EGGVIVRVDPDGR--T
Mflv_4759|M.gilvum_PYR-GCK THVA-SDPHVRTNDGGCDPTGGFVIGTMAYDERTGGGAVYRVAPDCH---
MSMEG_6378|M.smegmatis_MC2_155 LELERDLPSNRLNDGKVDKRGRFVFGSMDTLEESRSGKLYSYGPDGT---
.: .* * * .* * : .*
MMAR_0391|M.marinum_M AQVVAEDLDFPNGMVITPDHDTLIVAESVGRRLSAFTVSAD-GALNDRRV
MAV_5132|M.avium_104 AKVVADDLDFPNGMAITPDGATLIVAESTGRRLSAFAIDDD-GALSGRRV
TH_0278|M.thermoresistible__bu VTVVAEDLDFPNGMVITEDRSTLIVAESIGRRLTSYRIAAD-GTLTDREV
Mvan_2237|M.vanbaalenii_PYR-1 CEVVAEDLQFPNGMAITADGATLIVAESTGRRLTAFTIAED-GSLSGRRV
Mflv_4759|M.gilvum_PYR-GCK ATTLLAPVSISNGIQWSADGTRVFYIDTPTRRIDVFDVDLDTGRWSNRRV
MSMEG_6378|M.smegmatis_MC2_155 LQILDEDITVSNGPCWSPDGRTLYFCDTWTGEIWCYDYDLDTGKVSNRRT
: : ..** : * : :: .: : * * ..*..
MMAR_0391|M.marinum_M FAAGLD---GPPDGIALDAEGAVWAAMTLAHQFERIVAGGVVTDRIDMGD
MAV_5132|M.avium_104 FADGLD---GPPDGIALDADGGVWAAMTLAHQFERIVAGGAVTDRIDIGD
TH_0278|M.thermoresistible__bu FADGLD---GPPDGIALDAAGAVWTSMTLAHRFDRILPGGEVTDRIDIGG
Mvan_2237|M.vanbaalenii_PYR-1 FADGLD---GPPDGICLDAAGGVWVAMTLAHQFERIEEGGEVTDRIDIGA
Mflv_4759|M.gilvum_PYR-GCK HIDMNNSI-GYPDGMAIDVDQGLWVAMWGGGAVNHYDAFGQLVETIEVPG
MSMEG_6378|M.smegmatis_MC2_155 FARVDTSGGGAADGATVDAEGYLWQALVYGGKIIRYAPDGTVDHVIDFPV
. * .** :*. :* :: . . : * : . *:.
MMAR_0391|M.marinum_M RVAIA-CALGGPGRRTLFLLSSTDAYPKRLIGT---RLSQLDAVTVATPG
MAV_5132|M.avium_104 RVAIA-CALGGPQRRTLFLLSSTDAYPQRLVGT---RLSRLDAVTVTTPG
TH_0278|M.thermoresistible__bu RAAIA-CALGGPGRNLLFLLSTTDAYPKRLIGT---QSARLDVVKVDVAG
Mvan_2237|M.vanbaalenii_PYR-1 RTAIA-CTLGGPEDRTLFMLTAADAYPERLRGT---TLSRLDATTVDIPA
Mflv_4759|M.gilvum_PYR-GCK VTQVSSCAFGGGSLDELFITTSRQGLPNDSEPS---AGAIFSTKTTTRGA
MSMEG_6378|M.smegmatis_MC2_155 LKPTS-VMFGGPDLDVLYVTSMARPPLPRFPEDGQLRGSLFAIHGLGVRG
: :** *:: : : : .
MMAR_0391|M.marinum_M AGLP----
MAV_5132|M.avium_104 AGLP----
TH_0278|M.thermoresistible__bu AGLP----
Mvan_2237|M.vanbaalenii_PYR-1 ASPA----
Mflv_4759|M.gilvum_PYR-GCK ALLEFAG-
MSMEG_6378|M.smegmatis_MC2_155 VPEPRFGA
.