For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MRCLIVDDSASFRTAAATMLDSGGVEVVGVASNSAEAMVFCRELQPDVVLVDIALGAESGFELTEKLLGD LEPRPPAVILISTYDEQDLSEMIASSPAVGFVPKFALSVPAIQNLLATI
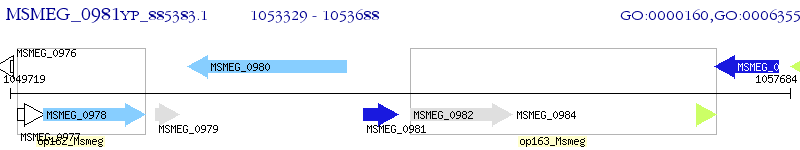
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0981 | - | - | 100% (119) | two-component system regulator |
| M. smegmatis MC2 155 | MSMEG_2251 | - | 3e-27 | 56.78% (118) | nitrate/nitrite response regulator protein |
| M. smegmatis MC2 155 | MSMEG_4378 | - | 8e-10 | 28.07% (114) | two-component system response regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1652 | - | 5e-09 | 27.83% (115) | two-component system transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_3575 | - | 3e-09 | 26.96% (115) | response regulator receiver/ANTAR domain-containing protein |
| M. tuberculosis H37Rv | Rv1626 | - | 5e-09 | 27.83% (115) | two-component system transcriptional regulator |
| M. leprae Br4923 | MLBr_01286 | - | 4e-09 | 27.19% (114) | putative two-component response regulatory protein |
| M. abscessus ATCC 19977 | MAB_2627c | - | 3e-09 | 27.83% (115) | two-component response regulatory protein |
| M. marinum M | MMAR_2429 | - | 3e-08 | 27.83% (115) | two-component system transcriptional regulator |
| M. avium 104 | MAV_3158 | - | 3e-09 | 27.83% (115) | response regulator |
| M. thermoresistible (build 8) | TH_1079 | - | 3e-09 | 27.43% (113) | Probable two-component system transcriptional regulator |
| M. ulcerans Agy99 | MUL_1608 | - | 2e-08 | 27.83% (115) | two-component system transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_2072 | - | 2e-29 | 57.39% (115) | response regulator receiver protein |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2429|M.marinum_M ------------------MTGPTTDADAAKTRRVLIAEDEALIRMDLAEM
MUL_1608|M.ulcerans_Agy99 ---------------MSVMTGPTTDADAAKTRRVLIAEDEALIRMDLAEM
Mb1652|M.bovis_AF2122/97 ------------------MTGPTTDADAAVPRRVLIAEDEALIRMDLAEM
Rv1626|M.tuberculosis_H37Rv ------------------MTGPTTDADAAVPRRVLIAEDEALIRMDLAEM
MAV_3158|M.avium_104 ---------------MSVMTGPTTDTDAAVPRRVLIAEDEALIRLDLAEM
MLBr_01286|M.leprae_Br4923 ------------------MTGPTTDADAATPHRVLIAEDEALIRLDLAEM
Mflv_3575|M.gilvum_PYR-GCK MAPRSPISPAGFTMFSMSSASASPSDAAATPHRVLIAEDEALIRMDLAEM
TH_1079|M.thermoresistible__bu ---------------------------------VLVAEDEALIRLDLAEM
MAB_2627c|M.abscessus_ATCC_199 ------------------MTGPQNTEPAAPARRVLVAEDEALIRLDLVEM
MSMEG_0981|M.smegmatis_MC2_155 -------------------------------MRCLIVDDSASFRTAAATM
Mvan_2072|M.vanbaalenii_PYR-1 ------MVVPNFPVNRNANGLRSCQARRMNGPRCLIVDDSADFRDAARAM
*:.:*.* :* *
MMAR_2429|M.marinum_M LREEGYDIVGEAGDGQEAVELAELHKPDLVIMDVKMPRRDGIDAASEIAT
MUL_1608|M.ulcerans_Agy99 LREEGYDIVGEAGDGQEAVELAELHKPDLVIMDVKMPRRDGIDAASEIAT
Mb1652|M.bovis_AF2122/97 LREEGYEIVGEAGDGQEAVELAELHKPDLVIMDVKMPRRDGIDAASEIAS
Rv1626|M.tuberculosis_H37Rv LREEGYEIVGEAGDGQEAVELAELHKPDLVIMDVKMPRRDGIDAASEIAS
MAV_3158|M.avium_104 LREEGYEIVGEAGDGQEAVELAERHRPDLVIMDVKMPRRDGIDAASEIAS
MLBr_01286|M.leprae_Br4923 LRDEGYDIVGEAADGQEAVELAERHKPDLVIMDVKMPRRDGIDAASEIAS
Mflv_3575|M.gilvum_PYR-GCK LREEGYEIVGEAGDGQEAVDLAESLRPDLVIMDVKMPRRDGIDAASEIAS
TH_1079|M.thermoresistible__bu LRDEGYDVVGEAGDGQEAVELAESLRPDLVIMDVKMPRRDGIDAAREIAA
MAB_2627c|M.abscessus_ATCC_199 LHEEGYDVVGQAGDGQEAVDLAEELKPDLVIMDVKMPRRDGIDAASEIAT
MSMEG_0981|M.smegmatis_MC2_155 LDSGGVEVVGVASNSAEAMVFCRELQPDVVLVDIALGAESGFELTEKLLG
Mvan_2072|M.vanbaalenii_PYR-1 LERGGIVVVGTARDAAEALRRAQELHPDVALVDVDLGAESGFDVAER---
* * :** * :. **: .. :**:.::*: : ..*:: : .
MMAR_2429|M.marinum_M KRIAPIVVLTAFSQRDLVERARDAGAMAYLVKPFTVSDLVPAIELAVSRF
MUL_1608|M.ulcerans_Agy99 KRIAPIVVLTAFSQRDLVERARDAGAMAYLVKPFTVSDLVPAIELAVSRF
Mb1652|M.bovis_AF2122/97 KRIAPIVVLTAFSQRDLVERARDAGAMAYLVKPFSISDLIPAIELAVSRF
Rv1626|M.tuberculosis_H37Rv KRIAPIVVLTAFSQRDLVERARDAGAMAYLVKPFSISDLIPAIELAVSRF
MAV_3158|M.avium_104 KRIAPIVVLTAFSQRDLVERARDAGAMAYLVKPFTISDLIPAIELAVSRF
MLBr_01286|M.leprae_Br4923 KRIAPIVVLTAFSQRDLVERARDAGAMAYLVKPFTISDLIPAIALAMSRF
Mflv_3575|M.gilvum_PYR-GCK KRIAPIVILTAFSQRELVERARDAGAMAYLVKPFSITDLIPAIEVAVSRF
TH_1079|M.thermoresistible__bu KRIAPVVVLTAFSQRDLVEQARDAGAMAYLVKPFSITDLIPAIEVAVSRF
MAB_2627c|M.abscessus_ATCC_199 KRIAPIVILTAFSQRDLVEKARDAGAMAYLVKPFSKTDLVPAIEVAVSRF
MSMEG_0981|M.smegmatis_MC2_155 D-------LEPRPPAVILISTYDEQDLSEMIASSPAVGFVPKFALSVP--
Mvan_2072|M.vanbaalenii_PYR-1 --------LDDVP--VILTSTHDEQDFADLIAASPALGFLPKLTLSPA--
* . :: : * :: :: . . .::* : :: .
MMAR_2429|M.marinum_M SEISALENEVATLSDRLETRKLVERAKGLLQAKQGMTEPEAFKWIQRAAM
MUL_1608|M.ulcerans_Agy99 SEISALENEVATLSDRLETRKLVERAKGLLQAKQGMTEPEAFKWIQRAAM
Mb1652|M.bovis_AF2122/97 REITALEGEVATLSERLETRKLVERAKGLLQTKHGMTEPDAFKWIQRAAM
Rv1626|M.tuberculosis_H37Rv REITALEGEVATLSERLETRKLVERAKGLLQTKHGMTEPDAFKWIQRAAM
MAV_3158|M.avium_104 SEIAELEREVATLSERLETRKLVERAKGLLQTEQGMTEPEAFKWIQRAAM
MLBr_01286|M.leprae_Br4923 SELTALEREVATLADRLETRKLVERAKGLLQVKQGMTEPEAFKWIQRAAM
Mflv_3575|M.gilvum_PYR-GCK SEIAELEKEVATLSDRLETRKLVERAKGLLQAKQGMTEPEAFKWIQRAAM
TH_1079|M.thermoresistible__bu AEITALESEVAELSERLETRKLVERAKGLLQAKHNMTEPEAFKWIQRAAM
MAB_2627c|M.abscessus_ATCC_199 GELTALEKEVQTLADRLETRKLVERAKGVLQSTQGLTEPEAFKWIQRAAM
MSMEG_0981|M.smegmatis_MC2_155 -----------------AIQNLLATI------------------------
Mvan_2072|M.vanbaalenii_PYR-1 -----------------AVRKLLDRVSGSPGT------------------
::*:
MMAR_2429|M.marinum_M DRRTTMKRVAEVVLETLDTPDEA------
MUL_1608|M.ulcerans_Agy99 DRRTTMKRVAEVVLETLDTPDEA------
Mb1652|M.bovis_AF2122/97 DRRTTMKRVAEVVLETLGTPKDT------
Rv1626|M.tuberculosis_H37Rv DRRTTMKRVAEVVLETLGTPKDT------
MAV_3158|M.avium_104 DRRTTMKRVAEVVLETLGTPKE-------
MLBr_01286|M.leprae_Br4923 DRRTTMKRVAEVVLETLDTPKDT------
Mflv_3575|M.gilvum_PYR-GCK DRRTTMKRVAEVVLETLDPPAASPEPEAT
TH_1079|M.thermoresistible__bu DRRTTMKRVAEVVLENLDTPKDQTPSN--
MAB_2627c|M.abscessus_ATCC_199 DRRTTMKRVAEVVLETLDTDAPS------
MSMEG_0981|M.smegmatis_MC2_155 -----------------------------
Mvan_2072|M.vanbaalenii_PYR-1 -----------------------------