For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTIAEIAGRTLTQERRLVTAIPGPISQELQARKQSAVAAGVGVTLPVYVVAAGGGVLADADGNQLIDFGS GIAVTTVGNSAPAVVDAVTQQVAAFTHTCFMVTPYEGYVKVAEHLNRLTPGDHEKRTALFNSGAEAVENA VKIARAYTRRQAVVVFDHAYHGRTNLTMAMTAKNQPYKHGFGPFANEVYRVPTSYPFRDGETDGAAAAAH ALDLINKQVGADNVAAVVIEPVHGEGGFVVPAPGFLGALQKWCTDNGAVFVADEVQTGFARTGALFACEH ENVVPDLIVTAKGIAGGLPLSAVTGRAEIMDGPQSGGLGGTYGGNPLACAAALAVIDTIERENLVARARA IGETMLSRLGALAAADPRIGEVRGRGAMIAVELVKPGTTEPDADLTKRVAAAAHAQGLVVLTCGTYGNVL RFLPPLSMPDHLLDEGLDILAAVFAEVK
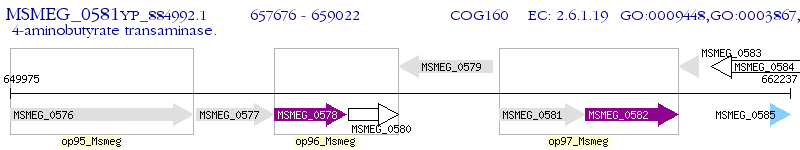
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0581 | gabT | - | 100% (448) | 4-aminobutyrate transaminase |
| M. smegmatis MC2 155 | MSMEG_2959 | gabT | e-180 | 72.08% (437) | 4-aminobutyrate aminotransferase |
| M. smegmatis MC2 155 | MSMEG_6685 | gabT | e-156 | 60.94% (448) | 4-aminobutyrate transaminase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2620 | gabT | 1e-176 | 68.68% (447) | 4-aminobutyrate aminotransferase |
| M. gilvum PYR-GCK | Mflv_3818 | - | 1e-178 | 69.35% (447) | 4-aminobutyrate aminotransferase |
| M. tuberculosis H37Rv | Rv2589 | gabT | 1e-176 | 68.68% (447) | 4-aminobutyrate aminotransferase |
| M. leprae Br4923 | MLBr_00485 | gabT | 1e-173 | 65.91% (443) | 4-aminobutyrate aminotransferase |
| M. abscessus ATCC 19977 | MAB_4207 | - | 1e-162 | 64.75% (434) | 4-aminobutyrate aminotransferase |
| M. marinum M | MMAR_2118 | gabT | 0.0 | 69.82% (444) | 4-aminobutyrate aminotransferase GabT |
| M. avium 104 | MAV_3470 | gabT | 1e-170 | 66.14% (446) | 4-aminobutyrate aminotransferase |
| M. thermoresistible (build 8) | TH_3529 | - | 2e-53 | 35.71% (406) | PUTATIVE aminotransferase |
| M. ulcerans Agy99 | MUL_1723 | gabT | 1e-179 | 69.37% (444) | 4-aminobutyrate aminotransferase |
| M. vanbaalenii PYR-1 | Mvan_2581 | - | 0.0 | 70.43% (443) | 4-aminobutyrate aminotransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2620|M.bovis_AF2122/97 -------MASLQQSRRLVTEIPGPASQALTHRRAAAVSSGVGVTLPVFVA
Rv2589|M.tuberculosis_H37Rv -------MASLQQSRRLVTEIPGPASQALTHRRAAAVSSGVGVTLPVFVA
MLBr_00485|M.leprae_Br4923 -------MTSVEQSRQLVTEIPGPVSLELAKRLNAAVPRGVGVTLPVFVT
MAV_3470|M.avium_104 -------MARLEQTRRLVTEIPGPASLELSKRRAAAVSHAVNVSVPVFVA
MMAR_2118|M.marinum_M -------MASLDQSRLLVTEIPGPASLELNKRRAAAVSGGVGVTLPVFVV
MUL_1723|M.ulcerans_Agy99 -------MASLDQSRLLVTEIPGPASLELNKRRAAAVSGGVGVTLPVFVV
Mflv_3818|M.gilvum_PYR-GCK -------MSALEQSRHLATAIPGPRSVELASRKNTAVSRGVGTTMPVYAA
Mvan_2581|M.vanbaalenii_PYR-1 -------MSALEQSRHLATAIPGPRSCELIARKSAAVARGVGTTMPVYAA
MSMEG_0581|M.smegmatis_MC2_155 MTIAEIAGRTLTQERRLVTAIPGPISQELQARKQSAVAAGVGVTLPVYVV
MAB_4207|M.abscessus_ATCC_1997 ---MTDITYRLAQKRTIVTPLPGPRSGALAERRRAAVSAGVGSTAPVYAV
TH_3529|M.thermoresistible__bu --------------------VSGSKGRHTLLDRWQGVMMNNYGTPPLALV
:.*. . .* : *: .
Mb2620|M.bovis_AF2122/97 RAGGGIVEDVDGNRLIDLGSGIAVTTIGNSSPRVVDAVRTQVAEFTHTCF
Rv2589|M.tuberculosis_H37Rv RAGGGIVEDVDGNRLIDLGSGIAVTTIGNSSPRVVDAVRTQVAEFTHTCF
MLBr_00485|M.leprae_Br4923 RAAGGVIEDVDGNRLIDLGSGIAVTTIGNSSPRVVDAVRAQVADFTHTCF
MAV_3470|M.avium_104 RAGGGIVEDVDGNRLIDLGSGIAVTTIGNSAPRVVDAVRAQVAEFTHTCF
MMAR_2118|M.marinum_M RAGGGIVEDADGNRLIDLGSGIAVTTIGNSAPRVVDAVRDQVEQFTHTCF
MUL_1723|M.ulcerans_Agy99 RAGGGIVEDADGNRLIDLGSGIAVTTIGNSAPRVVDAVRDQVEQFTHTCF
Mflv_3818|M.gilvum_PYR-GCK RAFGGIVEDVDGNRLIDLGSGIAVTTVGNASPRVVAAVAEQAAEFTHTCF
Mvan_2581|M.vanbaalenii_PYR-1 RAFGGIVEDVDGNRLIDLGSGIAVTTIGNASPKVVERAAEQLHKFTHTCF
MSMEG_0581|M.smegmatis_MC2_155 AAGGGVLADADGNQLIDFGSGIAVTTVGNSAPAVVDAVTQQVAAFTHTCF
MAB_4207|M.abscessus_ATCC_1997 DADGGVIVDADGNSFIDLGAGIAVTTVGASHPAVAAAIADQATHFTHTCF
TH_3529|M.thermoresistible__bu SGDGAVVTDADGKSYLDLLGGIAVNILGHRHPAVIEAVTRQLNTLGHTSN
. *.:: *.**: :*: .****. :* * * * : **.
Mb2620|M.bovis_AF2122/97 MVTPYEGYVAVAEQLNRITPGSGPKRSVLFNSGAEAVENAVKIARSYTGK
Rv2589|M.tuberculosis_H37Rv MVTPYEGYVAVAEQLNRITPGSGPKRSVLFNSGAEAVENAVKIARSYTGK
MLBr_00485|M.leprae_Br4923 IITPYEEYVAVTEQLNRITPGSGEKRSVLFNSGAEAVENSIKVARSYTRK
MAV_3470|M.avium_104 MVTPYESYVAVAETLNRITPGAGEKRSVLFNSGAEAVENAVKVARAYTRK
MMAR_2118|M.marinum_M MVTPYEQYVAVAEQLNRITPGSGEKRTVLFNSGAEAVENSIKVARAHTRK
MUL_1723|M.ulcerans_Agy99 MVIPYEHYVAVAEQLNRITPGSGEKRTVLFNSGAEAVENSIKVARAHTRK
Mflv_3818|M.gilvum_PYR-GCK MVTPYEGYVAVAEALNRLTPGDGDKRSALFNSGSEAVENAVKIARTFTRK
Mvan_2581|M.vanbaalenii_PYR-1 MVTPYEGYVAVAEALNRLTPGDGDKRTALFNSGSEAVENAVKIARSYTRK
MSMEG_0581|M.smegmatis_MC2_155 MVTPYEGYVKVAEHLNRLTPGDHEKRTALFNSGAEAVENAVKIARAYTRR
MAB_4207|M.abscessus_ATCC_1997 MVTPYEGYVQVAELLNALTPGDHDKRTALFNSGAEAVENAIKVARLATGR
TH_3529|M.thermoresistible__bu LYATEPGVALAEALVGHLG---APARVFFCNSGAEANEVAFKISR-LTGR
: . . . :. : * : ***:** * :.*::* * :
Mb2620|M.bovis_AF2122/97 PAVVAFDHAYHGRTNLTMALTAKSMPYKSGFGPFAPEIYRAPLSYPYRDG
Rv2589|M.tuberculosis_H37Rv PAVVAFDHAYHGRTNLTMALTAKSMPYKSGFGPFAPEIYRAPLSYPYRDG
MLBr_00485|M.leprae_Br4923 PAVVAFDHAYHGRTNLTMALTAKSMPYKSGFGPFAPEIYRAPLSYPYRDG
MAV_3470|M.avium_104 PAVVAFDHAYHGRTNLAMALTAKSMPYKGGFGPFAPEIYRAPLSYPYRDG
MMAR_2118|M.marinum_M QAVVAFDYAYHGRTNLTMALTAKSMPYKSGFGPFAPEIYRAPVSYPYRDN
MUL_1723|M.ulcerans_Agy99 QAVVAFDYAYHGRTNLTMALTAKSMPYKSGFGPFAPEIYRAPVSYPYRDN
Mflv_3818|M.gilvum_PYR-GCK QAVVSFDHAYHGRTNLTMALTAKSMPYKHGFGPFAPEIYRAPMSYPFRDA
Mvan_2581|M.vanbaalenii_PYR-1 QAVVSFDHAYHGRTNLTMALTAKSMPYKHGFGPFAPEIYRAPLSYPFRDA
MSMEG_0581|M.smegmatis_MC2_155 QAVVVFDHAYHGRTNLTMAMTAKNQPYKHGFGPFANEVYRVPTSYPFRD-
MAB_4207|M.abscessus_ATCC_1997 PAVVAFDNAYHGRTNLTMALTAKSMPYKSQFGPFAPEVYRMPASYPLRD-
TH_3529|M.thermoresistible__bu TKVVAAEGGFHGRTMGALALTGQP-SKREPFEPLPGDVIHVPYG------
** : .:**** ::*:*.: . : * *:. :: : * .
Mb2620|M.bovis_AF2122/97 LLD-KQLATNGELAAARAIGVIDKQVGANNLAALVIEPIQGEGGFIVPAE
Rv2589|M.tuberculosis_H37Rv LLD-KQLATNGELAAARAIGVIDKQVGANNLAALVIEPIQGEGGFIVPAE
MLBr_00485|M.leprae_Br4923 LLN-KDLATDGKLAGARAINVIEKQVGADDLAAVIIEPIQGEGGFIVPAE
MAV_3470|M.avium_104 LLD-KELATDGEKAAARAINIIDGQIGADSLAAVIIEPIQGEGGFIVPAE
MMAR_2118|M.marinum_M LLD-KDIATDGELAAERAINLIDKQIGAANLAAVIIEPIAGEGGFIVPAD
MUL_1723|M.ulcerans_Agy99 LLD-KDIATDGELAAERAINLIDKQIGAANLAAVIIEPIAGEGGFIVPAD
Mflv_3818|M.gilvum_PYR-GCK EFGGKELATDGELAARRAITVIDKQVGAGNLAAVIIEPIQGEGGFIVPAE
Mvan_2581|M.vanbaalenii_PYR-1 EFG-KELATDGELAARRAITVIDKQVGADNLAAVIIEPIQGEGGFIVPAE
MSMEG_0581|M.smegmatis_MC2_155 --G----ETDGAAAAAHALDLINKQVGADNVAAVVIEPVHGEGGFVVPAP
MAB_4207|M.abscessus_ATCC_1997 -----EPGLTGEEAARRAISRIETQIGAQSLAAIIIEPIQGEGGFIVPAP
TH_3529|M.thermoresistible__bu ----------DAGALERAV--------DDTTAAVFLEPIMGEGGVVVPPA
. * :*: **:.:**: ****.:**.
Mb2620|M.bovis_AF2122/97 GFLPALLDWCRKNHVVFIADEVQTGFARTGAMFACEHEGPDGLEPDLICT
Rv2589|M.tuberculosis_H37Rv GFLPALLDWCRKNHVVFIADEVQTGFARTGAMFACEHEGPDGLEPDLICT
MLBr_00485|M.leprae_Br4923 GFLATLLDWCRKNNVMFIADEVQTGFARTGAMFACEHDG---IVPDLICT
MAV_3470|M.avium_104 GFLPTLLDWCRRNDVLFIADEVQTGFARSGAMFACEHEGPNGLEPDLICT
MMAR_2118|M.marinum_M GFLPALQRWCRDNDVVFIADEVQTGFARTGAMFACDHEN---VEPDLIVT
MUL_1723|M.ulcerans_Agy99 GFLPALQRWSRDNDVVFIADEVQTGFARTGAMFACDHEN---VEPDLIVT
Mflv_3818|M.gilvum_PYR-GCK GFLPTLLDWCRANDVVFIADEVQTGFARTGAMFACEHEGPDGLVPDLIVT
Mvan_2581|M.vanbaalenii_PYR-1 GFLPTLLEWCHANGVVFIADEVQTGFARTGAMFACEHEG---IVPDLIVT
MSMEG_0581|M.smegmatis_MC2_155 GFLGALQKWCTDNGAVFVADEVQTGFARTGALFACEHEN---VVPDLIVT
MAB_4207|M.abscessus_ATCC_1997 GFLATLTAWASENGVVFIADEVQTGFARTGAWFASEHEG---IVPDIVTM
TH_3529|M.thermoresistible__bu GYLAAAREVTARHGALLVLDEVQTGIGRTGTFFAHQHDG---ITPDVVTL
*:* : : .::: ******:.*:*: ** :*:. : **::
Mb2620|M.bovis_AF2122/97 AKGIADGLPLSAVTGRAEIMNAPHVGGLGGTFGGNPVACAAALATIATIE
Rv2589|M.tuberculosis_H37Rv AKGIADGLPLSAVTGRAEIMNAPHVGGLGGTFGGNPVACAAALATIATIE
MLBr_00485|M.leprae_Br4923 AKGIADGLPLAAVTGRAEIMNAPHVSGLGGTFGGNPVACAAALATITTIE
MAV_3470|M.avium_104 AKAIADGLPLSAVTGRAQIMDAPHVGGLGGTFGGNPLACAAALATIETIE
MMAR_2118|M.marinum_M AKGIADGFPLSAVTGRAEIMDAPHTSGLGGTFGGNPVACAAALATIETIE
MUL_1723|M.ulcerans_Agy99 AKGIADGFPLSAVTGRAEIMDAPHTSGLGGTFGGNPVACAAALATIETIE
Mflv_3818|M.gilvum_PYR-GCK AKGIAGGMPLSAVTGRADIMDAPHVSGLGGTYGGNPVACAAALATIETIE
Mvan_2581|M.vanbaalenii_PYR-1 AKGIADGMPLSAVTGRAEIMDAPHVSGLGGTYGGNPVACAAALATIETIE
MSMEG_0581|M.smegmatis_MC2_155 AKGIAGGLPLSAVTGRAEIMDGPQSGGLGGTYGGNPLACAAALAVIDTIE
MAB_4207|M.abscessus_ATCC_1997 AKGIAGGMPLSAVTGRAELMDAVYAGGLGGTYGGNPVTCAAAVAALGVMR
TH_3529|M.thermoresistible__bu AKGLGGGLPIGACLAVGPAGDLLTPGMHGSTFGGNPVCTAAAVAVLGALA
**.:..*:*:.* . . : . *.*:****: ***:*.: .:
Mb2620|M.bovis_AF2122/97 SDGLIERARQIERLVTDRLTTLQAVDDRIGDVRGRGAMIAVELVKSGTTE
Rv2589|M.tuberculosis_H37Rv SDGLIERARQIERLVTDRLTTLQAVDDRIGDVRGRGAMIAVELVKSGTTE
MLBr_00485|M.leprae_Br4923 NDGLIQRAQQIERLITDRLLRLQDADDRIGDVRGRGAMIAVELVKSGTAE
MAV_3470|M.avium_104 SDGLIERARQLERLITEPLLRLQARDDRIGDVRGRGAMIAIELVKSGTAE
MMAR_2118|M.marinum_M RDGMVERARQIERLVMDRLLRLQAADDRLGDVRGRGAMIAMELVKSGTAE
MUL_1723|M.ulcerans_Agy99 RDGMVERARQIERLVMDRLLRLQAADDRLGDVRGRGAMIAMELVKSGTAE
Mflv_3818|M.gilvum_PYR-GCK SDNLVARAAQIEALMKDRLGRMQAEDDRIGDVRGRGAMIAVELVKSGTLE
Mvan_2581|M.vanbaalenii_PYR-1 SDGLVARAAQIETLMKDKLGRLQAEDDRIGDVRGRGAMIAVELVKPGTLE
MSMEG_0581|M.smegmatis_MC2_155 RENLVARARAIGETMLSRLGALAAADPRIGEVRGRGAMIAVELVKPGTTE
MAB_4207|M.abscessus_ATCC_1997 ELDLPARARAIEASVTSRLSALAEEVDIIGEVRGRGAMLAIEIVKPGTLE
TH_3529|M.thermoresistible__bu EGDLINRADVLGKKLSHGIEELRHP--LVDHVRGRGLLCGVVLNAP----
.: ** : : : : :..***** : .: : .
Mb2620|M.bovis_AF2122/97 PDAGLTERLATAAHAAGVIILTCGMFGNIIRLLPPLTIGDELLSEGLDIV
Rv2589|M.tuberculosis_H37Rv PDAGLTERLATAAHAAGVIILTCGMFGNIIRLLPPLTIGDELLSEGLDIV
MLBr_00485|M.leprae_Br4923 PDPELTEKVATAAHATGVIILTCGMFGNIIRLLPPLTISDELLAEGLDIL
MAV_3470|M.avium_104 PDKDLTQRLSVAAHAAGVIVLTCGMFGNIIRLLPPLTISDELLAEGIDIL
MMAR_2118|M.marinum_M PDAALTQKLAAAAHAAGVIVLTCGMFGNVIRLLPPLTISDELLSEGLDIL
MUL_1723|M.ulcerans_Agy99 PDAALTQKLAAAAHAAGVIVLTCGMFGNVIRLLPPLTISDELLSEGLDIL
Mflv_3818|M.gilvum_PYR-GCK PDPELTRKLLTAAHAAGVIVLSCGTFGNVLRFLPPLAISDDLLLEGLDIL
Mvan_2581|M.vanbaalenii_PYR-1 PDPELTKNLLAAAHRAGVIVLSCGTFGNVLRFLPPLAISDELLLEGLDVL
MSMEG_0581|M.smegmatis_MC2_155 PDADLTKRVAAAAHAQGLVVLTCGTYGNVLRFLPPLSMPDHLLDEGLDIL
MAB_4207|M.abscessus_ATCC_1997 PDAALTKSIAAEALSQGVLILTCGTFGNVIRLLPPLVIGDDLLDEGITAL
TH_3529|M.thermoresistible__bu ------QAKAAEAAARDAGFLVNAAAPEVIRLAPPLIITETQIDEFLAAL
. . * . .* . :::*: *** : : : * : :
Mb2620|M.bovis_AF2122/97 CAILADL----
Rv2589|M.tuberculosis_H37Rv CAILADL----
MLBr_00485|M.leprae_Br4923 SRILGDF----
MAV_3470|M.avium_104 GGLLADL----
MMAR_2118|M.marinum_M CQILADL----
MUL_1723|M.ulcerans_Agy99 CQILADL----
Mflv_3818|M.gilvum_PYR-GCK ELILRDL----
Mvan_2581|M.vanbaalenii_PYR-1 ELILRDL----
MSMEG_0581|M.smegmatis_MC2_155 AAVFAEVK---
MAB_4207|M.abscessus_ATCC_1997 SDIIRAKASHQ
TH_3529|M.thermoresistible__bu PGVLDTAKEAS
::