For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSPPRLNLADFLCFSIYSANLAFGKAYKPLLDQRGITYTQYIAIVALYDQDHQTVSGLGEKLFLESNTLT PILKKLESLGYVTRRRDPDDERQVVVSLTEAGRELREQSFELELASATGLTVDEMRTMQKGIAKLRDNLR EHVRATR
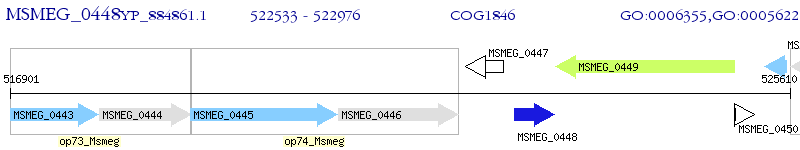
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0448 | - | - | 100% (147) | transcriptional regulator, MarR family protein |
| M. smegmatis MC2 155 | MSMEG_0538 | - | 5e-17 | 33.58% (134) | regulatory protein, MarR |
| M. smegmatis MC2 155 | MSMEG_5724 | - | 6e-07 | 26.45% (121) | transcriptional regulator MarR |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0043c | - | 1e-07 | 29.81% (104) | MarR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_0960 | - | 4e-26 | 38.89% (144) | MarR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv0042c | - | 1e-07 | 29.81% (104) | MarR family transcriptional regulator |
| M. leprae Br4923 | MLBr_02140 | - | 2e-05 | 28.77% (73) | putative MarR-family protein |
| M. abscessus ATCC 19977 | MAB_0925c | - | 9e-27 | 42.55% (141) | MarR family transcriptional regulator |
| M. marinum M | MMAR_0145 | - | 1e-17 | 40.94% (127) | transcriptional regulator |
| M. avium 104 | MAV_0057 | - | 2e-07 | 29.81% (104) | transcriptional regulator, MarR family protein |
| M. thermoresistible (build 8) | TH_3245 | - | 1e-07 | 31.82% (110) | PUTATIVE - |
| M. ulcerans Agy99 | MUL_4952 | - | 7e-18 | 40.94% (127) | transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_1524 | - | 4e-24 | 41.91% (136) | MarR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_0925c|M.abscessus_ATCC_199 -------------------------------------------------M
Mvan_1524|M.vanbaalenii_PYR-1 -------------------------------------------------M
Mflv_0960|M.gilvum_PYR-GCK -----------------------------------------------MRM
MMAR_0145|M.marinum_M -----------------------------------------MAAPTTPQA
MUL_4952|M.ulcerans_Agy99 -----------------------------------------MAAPTTPQA
MSMEG_0448|M.smegmatis_MC2_155 -----------------------------------------------MSP
Mb0043c|M.bovis_AF2122/97 MSVVRSIGKKMQRISGPNALAVKGRPTQVYGHTHVRLDCRFMADSEFTAP
Rv0042c|M.tuberculosis_H37Rv MSVVRSIGKKMQRISGPNALAVKGRPTQVYGHTHVRLDCRFMADSEFTAP
MAV_0057|M.avium_104 -----------------------------------------MADSESNAA
MLBr_02140|M.leprae_Br4923 -----------------------------------------MLDSD----
TH_3245|M.thermoresistible__bu --------------------------------------------MEERSS
MAB_0925c|M.abscessus_ATCC_199 SALGLDHQLCFALYSASRAMTAVYRPILTELNLTYPQYLVLLALWEEDRA
Mvan_1524|M.vanbaalenii_PYR-1 APLSLDEHLCFALYSASRAMTAAYRPLLTELGITYPQYLVLLVLGEEGDI
Mflv_0960|M.gilvum_PYR-GCK ARLRLDEQLCFALYSASRAVTSAYRPLLEELGLTYPQYLVLLVLWEEEPC
MMAR_0145|M.marinum_M DPLALEQQVCFALAITNRAVLAVYRPLLEPLGLTHPQYLVMLALWDNQRT
MUL_4952|M.ulcerans_Agy99 DPLALEQQVCFALAITNRAVLAVYRPLLEPLGLTHPQYLVMLALWDNQRT
MSMEG_0448|M.smegmatis_MC2_155 PRLNLADFLCFSIYSANLAFGKAYKPLLDQRGITYTQYIAIVALYDQDHQ
Mb0043c|M.bovis_AF2122/97 EVTQLAEGLHRALSKLISMLRRGDPNGAAAGDLTLAQLSILVTLLDQGPI
Rv0042c|M.tuberculosis_H37Rv EVTQLAEGLHRALSKLISMLRRGDPNGAAAGDLTLAQLSILVTLLDQGPI
MAV_0057|M.avium_104 EVAELAEGLHRALSKLFAMLRRGDPSGAAAGDLTLAQLSILVTLLDQGPI
MLBr_02140|M.leprae_Br4923 --ARLASDLSLAVMRLARQLRFRNPQSPVS----LSQLSALTTLANEGAM
TH_3245|M.thermoresistible__bu GIAALHQRIPFLLSQLGAHLAEDFVRRLEPFGIEPRIYAVLTALSEVDGQ
* : : . : .* :
MAB_0925c|M.abscessus_ATCC_199 TVS--------RLGERLRLDSGTLSPLLKRLETNGLIRRERSASDERLVE
Mvan_1524|M.vanbaalenii_PYR-1 SVG--------RLGERLQLDSGTLSPLLKRMEAGGIVRRQRSETDERQVE
Mflv_0960|M.gilvum_PYR-GCK TVG--------HLGERLHLDSGTLSPLLKRLEGAGLVQRQRSATDERRVT
MMAR_0145|M.marinum_M SSGQGSALAVKQIAATLQLDSATLSPMLKRLEALGLVTRTRNATDERSTD
MUL_4952|M.ulcerans_Agy99 SSGQGSALAVKQIAATLQLDSATLSPMLKRLEALGLVTRTRNATDERSTD
MSMEG_0448|M.smegmatis_MC2_155 TVS--------GLGEKLFLESNTLTPILKKLESLGYVTRRRDPDDERQVV
Mb0043c|M.bovis_AF2122/97 RMT--------DLAAHERVRTPTTTVAIRRLEKIGLVKRSRDPSDLRAVL
Rv0042c|M.tuberculosis_H37Rv RMT--------DLAAHERVRTPTTTVAIRRLEKIGLVKRSRDPSDLRAVL
MAV_0057|M.avium_104 RMT--------DLAAHERVRTPTTTVAIRRLEKVGLVKRSRDPSDLRAVL
MLBr_02140|M.leprae_Br4923 TPG--------ALAIRERVRPPSMTRVIASLADMGFVDREPHPVDGRQVL
TH_3245|M.thermoresistible__bu SQR--------ELSAQLGIHRNAMVAVVDKMESLGLVKRLPHPADRRAFA
:. : : : : * : * * *
MAB_0925c|M.abscessus_ATCC_199 VTLTPAGRHLERKAQCIPEKLALATG--IAEREATDLRDAVCQLTDALQA
Mvan_1524|M.vanbaalenii_PYR-1 VALTTEGKRLRRKVHCIPEKLFPLTG--LTPEQAIDLRADIVRLTEALID
Mflv_0960|M.gilvum_PYR-GCK ATLTPAGRALEERAACIPERMLGSTG--AAAEDLAALRDALHLITEELHT
MMAR_0145|M.marinum_M VKLTRFGIALRERAAEVPAAVIARLG--VDPTELEQLHRVLTRINVAALA
MUL_4952|M.ulcerans_Agy99 VKLTRFGIALRERAAEVPAAVIARLG--VDPTELEQLHRVLTRINVAALA
MSMEG_0448|M.smegmatis_MC2_155 VSLTEAGRELRE--QSFELELASATG--LTVDEMRTMQKGIAKLRDNLRE
Mb0043c|M.bovis_AF2122/97 VDITPQGRAVHGESLANRRAALAALLSQLPRSDLETLRKALAPLERLASG
Rv0042c|M.tuberculosis_H37Rv VDITPQGRAVHGESLANRRAALAALLSQLPRSDLETLRKALAPLERLASG
MAV_0057|M.avium_104 VDITPRGRAVHGESLANRRAALAAMLSQLPASDLDTLMKALAPLERLASG
MLBr_02140|M.leprae_Br4923 VSVSQSGAELVKAARRARQEWLVERLMTLNSDKRDILRNAADLILELIDE
TH_3245|M.thermoresistible__bu VTLTDKARELLPALDAEGRAQEDAVTAAMTPAERRETLRLLQKMSSTVGL
. :: . : . :
MAB_0925c|M.abscessus_ATCC_199 SLRKESA---------
Mvan_1524|M.vanbaalenii_PYR-1 SQEGNPHG--------
Mflv_0960|M.gilvum_PYR-GCK RSS-------------
MMAR_0145|M.marinum_M ADAPQS----------
MUL_4952|M.ulcerans_Agy99 ADAPQS----------
MSMEG_0448|M.smegmatis_MC2_155 HVRATR----------
Mb0043c|M.bovis_AF2122/97 EPASGPASNSPARKRA
Rv0042c|M.tuberculosis_H37Rv EPASGPASNSPARKRA
MAV_0057|M.avium_104 DPTSGPAGVPPARKEA
MLBr_02140|M.leprae_Br4923 SP--------------
TH_3245|M.thermoresistible__bu VPGVHPKLG-------