For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
STVHPTVAEVTRRIVERSAATRGVYLERMAEQTHRGPARGRLACANLAHGFAATEPADKEALRRIVKPNL AIVSSYNDMLSAHKPFEDYPAQLKAAALGAGAIAQVAGGVPAMCDGITQGRAGMQLSLFSRDVIAMSTAI ALSHDMFDAALLLGVCDKIVPGMLIGALSFGHLPAIFVPAGPMTSGLPNGEKSRIRQMFAEGKVDREALL DAEAASYHGRGTCTFYGTANSNQLLMEAMGLHLPGSSLVTPDTPLREALTAAAGARAAEVTALGDAYTPV GAVVDERAVVNGAVALLATGGSTNHTMHLVAIARAAGIILTWNDIADLSAVVPLLARIYPNGKADVNHFQ AAGGMAFLVRSLLDAGLLRDDVATVAGPGLRRYTTEPYLDDGALRWRDGVHASLDTDVLRGVDDPFAPDG GLKTLSGNLGTSVMKTSAVDPEHRTVTAPAVVFDSQEAFLEAFGAGELDRDFVGVLRYQGPQALGMPELH KLTPALGVLQDRGFKVALVTDGRMSGASGKVPAAIHVTPEAAAGGPLSRVRNGDVITVDATVGTLSVDVA PADLDARSSTGQAPTNDEWTGTGRELFATLRASVGPADEGATIFRSAV*
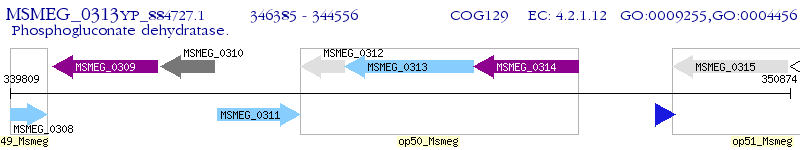
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0313 | edd | - | 100% (609) | phosphogluconate dehydratase |
| M. smegmatis MC2 155 | MSMEG_1290 | ilvD | 2e-63 | 32.88% (587) | dihydroxy-acid dehydratase |
| M. smegmatis MC2 155 | MSMEG_0229 | ilvD | 9e-57 | 31.08% (518) | dihydroxy-acid dehydratase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0195c | ilvD | 3e-56 | 30.38% (530) | dihydroxy-acid dehydratase |
| M. gilvum PYR-GCK | Mflv_5171 | - | 2e-58 | 33.33% (585) | dihydroxy-acid dehydratase |
| M. tuberculosis H37Rv | Rv0189c | ilvD | 3e-56 | 30.38% (530) | dihydroxy-acid dehydratase |
| M. leprae Br4923 | MLBr_02608 | ilvD | 2e-53 | 29.64% (533) | dihydroxy-acid dehydratase |
| M. abscessus ATCC 19977 | MAB_4640c | - | 1e-60 | 32.14% (585) | dihydroxy-acid dehydratase (IlvD) |
| M. marinum M | MMAR_0432 | ilvD | 3e-56 | 31.13% (530) | dihydroxy-acid dehydratase IlvD |
| M. avium 104 | MAV_4989 | ilvD | 2e-57 | 30.19% (530) | dihydroxy-acid dehydratase |
| M. thermoresistible (build 8) | TH_0983 | ilvD | 5e-58 | 31.65% (553) | PROBABLE DIHYDROXY-ACID DEHYDRATASE ILVD (DAD) |
| M. ulcerans Agy99 | MUL_1082 | ilvD | 3e-55 | 30.94% (530) | dihydroxy-acid dehydratase |
| M. vanbaalenii PYR-1 | Mvan_1161 | - | 7e-62 | 33.16% (588) | dihydroxy-acid dehydratase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0432|M.marinum_M ------MP-----------EADIKPRSRDVTDGLEKAAARGMLRAVGMVD
MUL_1082|M.ulcerans_Agy99 ------MP-----------EADIKPRSRDVTDGLEKAAARGMLRAVGMVD
MAV_4989|M.avium_104 ------MPTTDSARAADIKQPDIKPRSRDVTDGLEKAAARGMLRAVGMGD
Mb0195c|M.bovis_AF2122/97 ------MPQTTDEAASVSTVADIKPRSRDVTDGLEKAAARGMLRAVGMDD
Rv0189c|M.tuberculosis_H37Rv ------MPQTTDEAASVSTVADIKPRSRDVTDGLEKAAARGMLRAVGMDD
MLBr_02608|M.leprae_Br4923 -----MMP------------PDIKPRSRDVTDGLEKAAARGMLRAVGMND
TH_0983|M.thermoresistible__bu ------MAQ---------NQPDIKPRSRDVTDGLEKAAARGMLRAVGMGD
Mflv_5171|M.gilvum_PYR-GCK ---------------------MPELRSRTVTHGRNMAGARALLRAAGVDG
Mvan_1161|M.vanbaalenii_PYR-1 ----------------------MELRSRTVTHGRNMAGARSLLRAAGVDG
MAB_4640c|M.abscessus_ATCC_199 -----------------------------------MAGARALLRAAGVAA
MSMEG_0313|M.smegmatis_MC2_155 MSTVHPTVAEVTRRIVERSAATRGVYLERMAEQTHRGPARGRLACANLAH
. **. * ...:
MMAR_0432|M.marinum_M -------------EDFAKAQIGVASSWNEITPCNLSLDRLAKSVKEGVFA
MUL_1082|M.ulcerans_Agy99 -------------EDFAKAQIGVASSWNEITPCNLSLDRLAKSVKEGVFA
MAV_4989|M.avium_104 -------------EDFAKPQIGVASSWNEITPCNLSLDRLAKAVKEGVFA
Mb0195c|M.bovis_AF2122/97 -------------EDFAKPQIGVASSWNEITPCNLSLDRLANAVKEGVFS
Rv0189c|M.tuberculosis_H37Rv -------------EDFAKPQIGVASSWNEITPCNLSLDRLANAVKEGVFS
MLBr_02608|M.leprae_Br4923 -------------DDFAKAQIGVASSWNEITPCNLSLDRLAKAVKEGVFS
TH_0983|M.thermoresistible__bu -------------EDFAKPQIGVASSWNEITPCNLSLDRLAKAVKDGVHA
Mflv_5171|M.gilvum_PYR-GCK -------------ADIGKPIVAVANSFTEFVPGHTHLQPVGRIVSEAIRA
Mvan_1161|M.vanbaalenii_PYR-1 -------------ADIGKPIVAVANSFTEFVPGHTHLQPVGRIVSEAVTA
MAB_4640c|M.abscessus_ATCC_199 -------------SDFGKPIVAVANSFAEFVPGHTHLQPVGRIVSEAIAA
MSMEG_0313|M.smegmatis_MC2_155 GFAATEPADKEALRRIVKPNLAIVSSYNDMLSAHKPFEDYPAQLKAAALG
: *. :.:..*: :: . : :: :. . .
MMAR_0432|M.marinum_M AGGYPLEFG-TISVSDGISMGHEGMHFSLVSREVIADSVETVMQAERLDG
MUL_1082|M.ulcerans_Agy99 AGGYPLEFG-TISVSDGISMGHEGMHFSLVSREVIADSVETVMQAERLDG
MAV_4989|M.avium_104 AGGYPLEFG-TISVSDGISMGHEGMHFSLVSREVIADSVETVMQAERLDG
Mb0195c|M.bovis_AF2122/97 AGGYPLEFG-TISVSDGISMGHEGMHFSLVSREVIADSVEVVMQAERLDG
Rv0189c|M.tuberculosis_H37Rv AGGYPLEFG-TISVSDGISMGHEGMHFSLVSREVIADSVEVVMQAERLDG
MLBr_02608|M.leprae_Br4923 AGGYPLEFG-TISVSDGISMGHQGMHFSLVSREVIADSVETVMQAERLDG
TH_0983|M.thermoresistible__bu AGGYPLEFG-TISVSDGISMGHEGMHFSLVSREVIADSVETVMQAERLDG
Mflv_5171|M.gilvum_PYR-GCK AGGVPREFN-TIAVDDGIAMGHGGMLYSLPSRELIADSVEYMINAHCADA
Mvan_1161|M.vanbaalenii_PYR-1 AGGVPREFN-TIAVDDGIAMGHGGMLYSLPSRDLIADSIEYMVNAHRADA
MAB_4640c|M.abscessus_ATCC_199 AGAVPREFN-TIAVDDGIAMGHEGMLYSLPSRDLITDSIEYMVEAHRADA
MSMEG_0313|M.smegmatis_MC2_155 AGAIAQVAGGVPAMCDGITQGRAGMQLSLFSRDVIAMSTAIALSHDMFDA
**. . . . :: ***: *: ** ** **::*: * :. . *.
MMAR_0432|M.marinum_M SVLLAGCDKSLPGMLMAAARL-DLASVFLYAGSILPGVAKLSDGSEREVT
MUL_1082|M.ulcerans_Agy99 SVLLAGCDKSLPGMLMAAARL-DLASVFLYAGSILPGVAKLSDGSEREVT
MAV_4989|M.avium_104 SVLLAGCDKSLPGMLMAAARL-DLASVFLYAGSILPGVAKLSDGSEREVT
Mb0195c|M.bovis_AF2122/97 SVLLAGCDKSLPGMLMAAARL-DLAAVFLYAGSILPGRAKLSDGSERDVT
Rv0189c|M.tuberculosis_H37Rv SVLLAGCDKSLPGMLMAAARL-DLAAVFLYAGSILPGRAKLSDGSERDVT
MLBr_02608|M.leprae_Br4923 SVLLAGCDKSLPGMLMAAARL-DLASVFLYAGSILPGRTKLSDGTEHEVT
TH_0983|M.thermoresistible__bu SVLLAGCDKSLPGMLMAAARL-DLAAVFLYAGSILPGIAKLSDGTEREVT
Mflv_5171|M.gilvum_PYR-GCK MVCISNCDKITPGMVMAALRL-DIPTVFVSGGPMEGGSAVLVDGTVR--T
Mvan_1161|M.vanbaalenii_PYR-1 LVCISNCDKITPGMLMAALRL-NIPTVFVSGGPMEGGTAVLVDGTVR--T
MAB_4640c|M.abscessus_ATCC_199 LVCISNCDKITPGMLMAALRL-NIPAVFVSGGPMEGGRATLRDGTVR--T
MSMEG_0313|M.smegmatis_MC2_155 ALLLGVCDKIVPGMLIGALSFGHLPAIFVPAGPMTSGLPNGEKSRIR---
: :. *** ***::.* : .:.::*: .*.: * . .. :
MMAR_0432|M.marinum_M IIDAFEAVGACSRGLMSRADVDAIERAICPGEGACGGMYTANTMASAAEA
MUL_1082|M.ulcerans_Agy99 IIDAFEAVGACSRGLMSRADVDAIERAICPGEGACGGMYTANTMASAAEA
MAV_4989|M.avium_104 IIDAFEAVGACARGLMPREDVDAIERAICPGEGACGGMYTANTMASAAEA
Mb0195c|M.bovis_AF2122/97 IIDAFEAVGACSRGLMSRADVDAIERAICPGEGACGGMYTANTMASAAEA
Rv0189c|M.tuberculosis_H37Rv IIDAFEAVGACSRGLMSRADVDAIERAICPGEGACGGMYTANTMASAAEA
MLBr_02608|M.leprae_Br4923 LIDAFEAVGACSRGLMPRADVDAIERAICPGEGACGGMYTANTMASAAEA
TH_0983|M.thermoresistible__bu IIDAFEAVGACARGLMPREDVDAIERAICPGEGACGGMYTANTMASAAEA
Mflv_5171|M.gilvum_PYR-GCK RLNLVSAIADAVDSGVSDPDLARIEESACPTCGSCSGMFTANSMNCLTEA
Mvan_1161|M.vanbaalenii_PYR-1 RLNLISAIADAVDTTVSDRDLARIEESACPTCGSCSGMFTANSMNCLTEA
MAB_4640c|M.abscessus_ATCC_199 GMTLITAVAESASDSVSDEDLARIEDQACPTCGSCSGMFTANSMNCLTEA
MSMEG_0313|M.smegmatis_MC2_155 --------QMFAEGKVDREALLDAEAASYHGRGTCTFYGTANSNQLLMEA
: : * *:* ***: **
MMAR_0432|M.marinum_M LGMSLPGSAAPPATDRRRDGFARQSGQAVIELLRQ-------GITARDIM
MUL_1082|M.ulcerans_Agy99 LGMSLPGSAAPPATDRRRDGFARQSGQAVIELLRQ-------GITARDIM
MAV_4989|M.avium_104 LGMSLPGSAAPPATDRRRDGFARRSGQAVVELLRR-------GITARDIL
Mb0195c|M.bovis_AF2122/97 LGMSLPGSAAPPATDRRRDGFARRSGQAVVELLRR-------GITARDIL
Rv0189c|M.tuberculosis_H37Rv LGMSLPGSAAPPATDRRRDGFARRSGQAVVELLRR-------GITARDIL
MLBr_02608|M.leprae_Br4923 LGMSLPGSAAPPATDRRRDGFARRSGQAVIELLRR-------GITARDIL
TH_0983|M.thermoresistible__bu LGMSLPGSAAPPATDRRRDGFARKSGQAVVELLRR-------GITARDIL
Mflv_5171|M.gilvum_PYR-GCK LGLALPGNGSVLATHTARRALYENAGATVMDLCRRYYDEDDTSVLPRAIA
Mvan_1161|M.vanbaalenii_PYR-1 LGLALPGNGSVLATHTARRALYEDAGATVMDLCRRYYDEDDASVLPRAIA
MAB_4640c|M.abscessus_ATCC_199 LGLAPPGNGTTLATHTARRGLYEQAGSLIVELAQRYYGSDDTTVLPREIA
MSMEG_0313|M.smegmatis_MC2_155 MGLHLPGSSLVTPDTPLREALTAAAGARAAEVTALG----DAYTPVGAVV
:*: **.. . * .: :* :: :
MMAR_0432|M.marinum_M TREAFENAIAVVMAFGGSTNAVLHLLAIAHEANVELTLDDFSRIGSKVPH
MUL_1082|M.ulcerans_Agy99 TREAFENAIAVVMAFGGSTNAVLHLLAIAHEANVELTLDDFSRIGSKVPH
MAV_4989|M.avium_104 TKEAFENAIAVVMAFGGSTNAVLHLLAIAHEADVALSLDDFSRIGSKVPH
Mb0195c|M.bovis_AF2122/97 TKEAFENAIAVVMAFGGSTNAVLHLLAIAHEANVALSLQDFSRIGSGVPH
Rv0189c|M.tuberculosis_H37Rv TKEAFENAIAVVMAFGGSTNAVLHLLAIAHEANVALSLQDFSRIGSGVPH
MLBr_02608|M.leprae_Br4923 TKEAFENAIAVVMAFGGSTNAILHLLAIAHEADVTLSLEDFSRIGFKVPH
TH_0983|M.thermoresistible__bu TKEAFENAIAVVMAFGGSTNAVLHLLAIAHEANVKLTLEDFTRIGNKVPH
Mflv_5171|M.gilvum_PYR-GCK NREAFDNAMAMDMAMGGSTNTILHLLAAAREAELDYTLEDIEKRSRQIPC
Mvan_1161|M.vanbaalenii_PYR-1 NRDAFDNAMAMDIAMGGSTNTILHLLAAAREAELDYTLEDIEKRSRAVPC
MAB_4640c|M.abscessus_ATCC_199 SRRAFENAMCMDIAMGGSTNTVLHLLAAAHEAELDFALADIDALSRRVPC
MSMEG_0313|M.smegmatis_MC2_155 DERAVVNGAVALLATGGSTNHTMHLVAIARAAGIILTWNDIADLSAVVPL
. *. *. :* ***** :**:* *: * : : *: . :*
MMAR_0432|M.marinum_M LADVKPFGRHVMSHVDHIGGVPVVMKTLLDAGLLHGDCLTVTG-------
MUL_1082|M.ulcerans_Agy99 LADVKPFGRHVMSHVDHIGGVPVVMKTLLDAGLLHGDCLTVTG-------
MAV_4989|M.avium_104 LADVKPFGRHVMTDVDHIGGVPVMMKALLDAGLLNGDCLTVTG-------
Mb0195c|M.bovis_AF2122/97 LADVKPFGRHVMSDVDHIGGVPVVMKALLDAGLLHGDCLTVTG-------
Rv0189c|M.tuberculosis_H37Rv LADVKPFGRHVMSDVDHIGGVPVVMKALLDAGLLHGDCLTVTG-------
MLBr_02608|M.leprae_Br4923 IADVKPFGRHVMFDVDHIGGVPVVMKALLDAGLLHGDCLTVTG-------
TH_0983|M.thermoresistible__bu LADVKPFGAHVMFDVDHIGGVPVIMKTLLDAGLLHGDCLTVTG-------
Mflv_5171|M.gilvum_PYR-GCK LCKVAPNGHYLMEDVHRAGGIPAILGELWRGGHLHETVHSVHAGSLPEWL
Mvan_1161|M.vanbaalenii_PYR-1 LCKVAPNGHYLMEDVHRAGGIPAILGELWRGGHLHETVHSVHSRSLAQWL
MAB_4640c|M.abscessus_ATCC_199 LSKVAPNGAYLVEDVHRAGGVPALLGELNRAGLLHRDVHSVHAPDLTTWL
MSMEG_0313|M.smegmatis_MC2_155 LARIYPNGKADVNHFQAAGGMAFLVRSLLDAGLLRDDVATVAG-------
:. : * * : ... **:. :: * .* *. :* .
MMAR_0432|M.marinum_M ----------RTVAENLEAIAP-------------------PDPDGKVLR
MUL_1082|M.ulcerans_Agy99 ----------RTVAENLEAIAP-------------------PDPDGKVLR
MAV_4989|M.avium_104 ----------ATVAQNLAAIAP-------------------PDPDGKVLR
Mb0195c|M.bovis_AF2122/97 ----------HTMAENLAAITP-------------------PDPDGKVLR
Rv0189c|M.tuberculosis_H37Rv ----------HTMAENLAAITP-------------------PDPDGKVLR
MLBr_02608|M.leprae_Br4923 ----------QTMAENLASIAP-------------------PDPDGQVIR
TH_0983|M.thermoresistible__bu ----------KTMAENLAHIAP-------------------PDPDGKVLR
Mflv_5171|M.gilvum_PYR-GCK RRWDVRGGQACEEAIELFHAAPGCVRSASAFSQSERWESLDTDASTGCIR
Mvan_1161|M.vanbaalenii_PYR-1 RRWDIRGGSASPEAVELFHAAPGCVRSASAFSQSQRWDTLDTDAEEGCIR
MAB_4640c|M.abscessus_ATCC_199 TQWDIRGTAPSPEAIELFHAAPGGERSARAFSQSQRWHTLDLDAENGCIR
MSMEG_0313|M.smegmatis_MC2_155 -----PGLRRYTTEPYLDDGALRWRDG------------VHASLDTDVLR
* : . . :*
MMAR_0432|M.marinum_M ALNNPIHATGGITILRGSLAPEGAVVKTAGLDT--DVFEGTARVFDGERA
MUL_1082|M.ulcerans_Agy99 ALINPIHATGGITILRGSLAPEGAVVKTAGLDT--DVFEGTARVFDGERA
MAV_4989|M.avium_104 ALSDPLHPTGGITILRGSLAPEGAVVKSAGFDS--DVFEGTARVFDGERA
Mb0195c|M.bovis_AF2122/97 ALANPIHPSGGITILHGSLAPEGAVVKTAGFDS--DVFEGTARVFDGERA
Rv0189c|M.tuberculosis_H37Rv ALANPIHPSGGITILHGSLAPEGAVVKTAGFDS--DVFEGTARVFDGERA
MLBr_02608|M.leprae_Br4923 TLHNPIHPTGGITILRGSLAPDGAVVKTAGLDS--DVFEGTARVFDGERA
TH_0983|M.thermoresistible__bu ALSNPIHPTGGITILHGSLAPEGAVVKSAGFDT--SVFEGTARVFDRERA
Mflv_5171|M.gilvum_PYR-GCK DVRHAYSEDGGLAILRGNLSRDGCIVKTAGVDESIWTFSGPAVVVESQED
Mvan_1161|M.vanbaalenii_PYR-1 DVQHAYSADGGLAVLRGNLAVDGCIVKTAGVDESMLTFTGPAVVVESQEG
MAB_4640c|M.abscessus_ATCC_199 DTEHAYSADGGLAVLTGNLAPNGCIVKTAGVDEKIRVFSGPAVVLESQEA
MSMEG_0313|M.smegmatis_MC2_155 GVDDPFAPDGGLKTLSGNLG--TSVMKTSAVDPEHRTVTAPAVVFDSQEA
.. **: * *.*. .::*::..* .. ..* *.: :.
MMAR_0432|M.marinum_M ALDALEDGTITHGDVVVIRYEGPKGGPGMREMLAITGAIKGA-GLGKDVL
MUL_1082|M.ulcerans_Agy99 ALDALEDGTITHGDVVVIRYEGPKGGPGMREMLAITGAIKGA-GLGKDVL
MAV_4989|M.avium_104 ALDALEDGTITKGDAVVIRYEGPKGGPGMREMLAITGAIKGA-GLGKDVL
Mb0195c|M.bovis_AF2122/97 ALDALEDGTITVGDAVVIRYEGPKGGPGMREMLAITGAIKGA-GLGKDVL
Rv0189c|M.tuberculosis_H37Rv ALDALEDGTITVGDAVVIRYEGPKGGPGMREMLAITGAIKGA-GLGKDVL
MLBr_02608|M.leprae_Br4923 ALDALKDGTIAKGDVVVIRYEGPKGGPGMREMLAITGAIKGA-GLGKDVL
TH_0983|M.thermoresistible__bu AMDALEDGTIQAGDVVVIRYEGPKGGPGMREMLAITGAIKGA-GLGKDVL
Mflv_5171|M.gilvum_PYR-GCK AVDAILNGRVQPGDVVVVRFEGPKGGPGMQEMLYPTSYLKGR-GLGKVCA
Mvan_1161|M.vanbaalenii_PYR-1 AVEAILNGRARPGDVVVVRYEGPKGGPGMQEMLYPTSYLKGR-GLATVCA
MAB_4640c|M.abscessus_ATCC_199 AVEAILNDRVRPGDVVVIRYEGPRGGPGMQEMLYPTAFLKGR-GLGASCA
MSMEG_0313|M.smegmatis_MC2_155 FLEAFGAGELDRDFVGVLRYQGPQ-ALGMPELHKLTPALGVLQDRGFKVA
::*: . . . *:*::**: . ** *: * : . .
MMAR_0432|M.marinum_M LLTDGRFSGGT-TGFCVGHIAPEAVDAGPIAFVRDGDRIRLDVAGRTLDV
MUL_1082|M.ulcerans_Agy99 LLTDGRFSGGT-TGFCVGHIAPEAVDAGPIAFVRDGDRIRLDVAGRTLDV
MAV_4989|M.avium_104 LLTDGRFSGGT-TGLCVGHIAPEAVDAGPIAFLRDGDRIRLDVANRVLDV
Mb0195c|M.bovis_AF2122/97 LLTDGRFSGGT-TGLCVGHIAPEAVDGGPIALLRNGDRIRLDVAGRVLDV
Rv0189c|M.tuberculosis_H37Rv LLTDGRFSGGT-TGLCVGHIAPEAVDGGPIALLRNGDRIRLDVAGRVLDV
MLBr_02608|M.leprae_Br4923 LLTDGRFSGGT-TGFCVGHIAPEAVEAGPIAFLRDGDRVLLDVVGCSLDV
TH_0983|M.thermoresistible__bu LLTDGRFSGGT-TGLCVGHVAPEAVDGGPIAFLRDGDRIRLDVAKGTLDV
Mflv_5171|M.gilvum_PYR-GCK LVTDGRFSGGS-SGLAVGHVSPEAAAGGTIALVEDGDRITIDIPRRTIGL
Mvan_1161|M.vanbaalenii_PYR-1 LVTDGRFSGGS-SGLSIGHVSPEAAAGGVIALIEDGDTITIDIPHRTIAL
MAB_4640c|M.abscessus_ATCC_199 LITDGRFSGGT-SGLSIGHISPEAAAGGPIALVEDGDIINIDIARRSIVV
MSMEG_0313|M.smegmatis_MC2_155 LVTDGRMSGASGKVPAAIHVTPEAAAGGPLSRVRNGDVITVDATVGTLSV
*:****:**.: . . *::***. .* :: :.:** : :* : :
MMAR_0432|M.marinum_M LTDPAEFAARQQNFTP-------PAPRYKTGVLAKYVKLVGSAAIGAVCG
MUL_1082|M.ulcerans_Agy99 LTDPPEFAARQQNFTP-------PAPRYKTGVLAKYVKLVGSAAIGAVCG
MAV_4989|M.avium_104 LVDPAEFDSRRTAFTP-------PPPRYKTGVLAKYVKLVGSAAIGAVCG
Mb0195c|M.bovis_AF2122/97 LADPAEFASRQQDFSP-------PPPRYTTGVLSKYVKLVSSAAVGAVCG
Rv0189c|M.tuberculosis_H37Rv LADPAEFASRQQDFSP-------PPPRYTTGVLSKYVKLVSSAAVGAVCG
MLBr_02608|M.leprae_Br4923 LVDPVEFSSRKKDFIP-------PAPRYTTGVLAKYVKLVSSATVGAVCG
TH_0983|M.thermoresistible__bu LVDPAEFEARKKDWEP-------LPPRYTTGVLAKYTKLVGSAATGAVCT
Mflv_5171|M.gilvum_PYR-GCK DVADDELDRRRAALEG-GGYRPRHRERPVSAALRAYAALALSADKGAVRD
Mvan_1161|M.vanbaalenii_PYR-1 DVGDDELQRRRAALQASGGYRPRDRQRVVSAALRAYAALAMSADKGAVRD
MAB_4640c|M.abscessus_ATCC_199 AVDPQTLQRRRALLESGNGYRPVTRERHVSAALRAYAAMATSADKGAVRS
MSMEG_0313|M.smegmatis_MC2_155 DVAPADLDARSSTGQAPTNDEWTGTGRELFATLRASVGPADEGATIFRSA
. : * * ..* . . ..
MMAR_0432|M.marinum_M ----
MUL_1082|M.ulcerans_Agy99 ----
MAV_4989|M.avium_104 ----
Mb0195c|M.bovis_AF2122/97 ----
Rv0189c|M.tuberculosis_H37Rv ----
MLBr_02608|M.leprae_Br4923 ----
TH_0983|M.thermoresistible__bu ----
Mflv_5171|M.gilvum_PYR-GCK VGRS
Mvan_1161|M.vanbaalenii_PYR-1 VSAV
MAB_4640c|M.abscessus_ATCC_199 LSG-
MSMEG_0313|M.smegmatis_MC2_155 V---