For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VEPNGRDRLRELLDAVVDSENDGVGDMARNSYASEFHFSREVRRLTGEPPAALRRRIMLERAAWRLRQGA GVAEVAAAEGWSSPEVFSRAFRRAFGVPPSRATDVGFRLPAPNGVHFHPPRSLWCDGRAGGGGPDIAQFM LVHDIADTAYLIEQAYALTENQWRQEVSPGQTVLDWDGPEPSVAAVLAAIVWTKEVWLATIEGRDFPSRD TTCNASPRELAVHHDTVGNRWTTVISEYSTAGRLGDTVIDALCDPPESFQLYGIVAHVLTYSAHRRGLAR AMLSRHGIDTHRGDPLEWMRRD
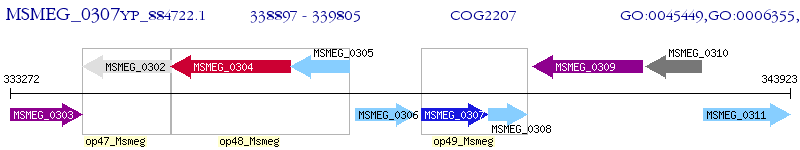
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0307 | - | - | 100% (302) | transcriptional regulator, AraC family protein |
| M. smegmatis MC2 155 | MSMEG_4655 | - | 2e-06 | 36.71% (79) | transcriptional regulator, AraC family protein |
| M. smegmatis MC2 155 | MSMEG_0376 | - | 2e-05 | 32.29% (96) | transcriptional regulator, AraC family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0432 | - | 1e-125 | 71.62% (303) | helix-turn-helix domain-containing protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0767c | - | 3e-08 | 37.97% (79) | AraC family transcriptional regulator |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_1906 | - | 1e-118 | 68.11% (301) | transcriptional regulator, AraC family protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_0236 | - | 1e-125 | 72.28% (303) | helix-turn-helix domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0432|M.gilvum_PYR-GCK -----------MNDP-----------------------------ARRDRL
Mvan_0236|M.vanbaalenii_PYR-1 MTDDPSPEHAALPDPSALRASAHPDPSALRASAHPDPSALRASAHRRDRL
MSMEG_0307|M.smegmatis_MC2_155 ------------MEP-----------------------------NGRDRL
TH_1906|M.thermoresistible__bu -----------VSEQ---------------------------ALTGRDRL
MAB_0767c|M.abscessus_ATCC_199 ----------MIHNP-------------------------DYFADQPKSL
: . *
Mflv_0432|M.gilvum_PYR-GCK RELLDAVTDAGNSGVGDMARSTYASEFHFSREVRRLTGEAPAALRRRVML
Mvan_0236|M.vanbaalenii_PYR-1 RELLDAVTDSANADVGDMARSSYASEFHFSREVRRLTGESPAALRRRVML
MSMEG_0307|M.smegmatis_MC2_155 RELLDAVVDSENDGVGDMARNSYASEFHFSREVRRLTGEPPAALRRRIML
TH_1906|M.thermoresistible__bu RELLDAVVDAANTDVESMARSSHFSEFHFSREVRRLAGEPPAAMRRRVML
MAB_0767c|M.abscessus_ATCC_199 SWPTGSPSIDAFSGILSAIGLRGESALRCSAELP-FDVSVPANERVLHIA
.: .: . * :: * *: : . ** * :
Mflv_0432|M.gilvum_PYR-GCK ERAAWRLQH-GEPVSAVAADEGWSSPEVFSRAFRRTYGVPPSQAVDVHFR
Mvan_0236|M.vanbaalenii_PYR-1 ERAAWRLQR-GEAVSAVAADEGWSSAEVFSRAFRRMYGLPPSQASDVHFR
MSMEG_0307|M.smegmatis_MC2_155 ERAAWRLRQ-GAGVAEVAAAEGWSSPEVFSRAFRRAFGVPPSRATDVGFR
TH_1906|M.thermoresistible__bu ERAAWRLQR-GESVSAVAAAEGWSSAEVFSRAFRRAFGVPPSRVAEVHYL
MAB_0767c|M.abscessus_ATCC_199 ETAGLRVYVDGNPVCVLAAGD--MVLMARGTAHRLSGAQPHGEPAGTLHW
* *. *: * *. :** : . . *.* . * .. . .
Mflv_0432|M.gilvum_PYR-GCK LPSPNGLHFHPPQSLWLDSDRSD--DPG--VPDIAALMIAHDIDDTAYLL
Mvan_0236|M.vanbaalenii_PYR-1 LPAPNGLHFHPPQSLWLDSDPGDRTPPGMHTPDIAQLMIDHDIADTSHLL
MSMEG_0307|M.smegmatis_MC2_155 LPAPNGVHFHPPRSLWCDGRAGG---GG---PDIAQFMLVHDIADTAYLI
TH_1906|M.thermoresistible__bu LPAPNGVHFHPPHALWVDGPGSRGAPAG-SVPEISAVLVDHDVADTDYLL
MAB_0767c|M.abscessus_ATCC_199 VSGRFTAERKAADPLLSVLPPAIIIQAA--RPGHEWLPLSLQLILAEILT
:.. . :.. .* . . * . : :: : *
Mflv_0432|M.gilvum_PYR-GCK TRAAELTRAQWTAEVAPGQTVLEWDGPEPSVGAVLGALVWNKEVWLATIA
Mvan_0236|M.vanbaalenii_PYR-1 GQAALLSPQQWVAEVAPGQTVLEWDGPEPSVGAVLGAIVWTKQVWLATIA
MSMEG_0307|M.smegmatis_MC2_155 EQAYALTENQWRQEVSPGQTVLDWDGPEPSVAAVLAAIVWTKEVWLATIE
TH_1906|M.thermoresistible__bu ECACTLTDRQWAEEISPGQQVLAWDGPEPSVGAVLGAIVWTKQVWLATIE
MAB_0767c|M.abscessus_ATCC_199 PSPGSWVMISRILDLLFIHAVREWSSSSHAEPGWLTAAMDPRLAPVLTAI
. . :: : * *.... : . * * : : . : *
Mflv_0432|M.gilvum_PYR-GCK GEDFPSRAATQPDLVDARTLAAHHDDIAKRWRAMISEYAAAGRLGDTVID
Mvan_0236|M.vanbaalenii_PYR-1 GDDFPSRACTQPEFVDAATLAAHHDDVGNRWRAMVSEYAVAGRLGDTVID
MSMEG_0307|M.smegmatis_MC2_155 GRDFPSRDTTCN--ASPRELAVHHDTVGNRWTTVISEYSTAGRLGDTVID
TH_1906|M.thermoresistible__bu GADFPTRQHLPP-----SALRDHHRDVGARWIEMVQSCSAAGKLGDTVID
MAB_0767c|M.abscessus_ATCC_199 HNNLEHPWSIAELAQHARQSRSAFTQRFTELLGQPPAAYIGERRLDRAAH
:: . . . : * . .
Mflv_0432|M.gilvum_PYR-GCK ALCDPPESFQLYGIVAHVLTYSAHRRELVRAMLAGHGVPTGLGDPLDWMR
Mvan_0236|M.vanbaalenii_PYR-1 ALCDPPESFQLYGIVAHVLTYSAHRRGLARTMLAGHGVTTRLGDPLDWMR
MSMEG_0307|M.smegmatis_MC2_155 ALCDPPESFQLYGIVAHVLTYSAHRRGLARAMLSRHGIDTHRGDPLEWMR
TH_1906|M.thermoresistible__bu ALCDPPESFPLYGIIAHVLTFSALRRGLARAMLAHHGVRTHHGDPLAWMR
MAB_0767c|M.abscessus_ATCC_199 LLRSTTDPVRQIGRAVGYTSDAAFSRAFQRRFGE---------PPLRWRT
* ...:.. * . : :* * : * : ** *
Mflv_0432|M.gilvum_PYR-GCK GR---
Mvan_0236|M.vanbaalenii_PYR-1 GN---
MSMEG_0307|M.smegmatis_MC2_155 RD---
TH_1906|M.thermoresistible__bu SN---
MAB_0767c|M.abscessus_ATCC_199 HHRSG