For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MVSSTRSEHSFAGVGGVRIVYDVWTPDTDPRGVVVLAHGYAEHAGRYHHVAQRFGAAGLLVYALDHRGHG RSGGKRVHLRDLSEFVEDFRTLVGIAANDHPTLPRIVLGHSMGGGIVFAYGARYPGEYSAMVLSGPAVNA HDGVSPVLVAVAKVLGKLAPGIPVENLDADAVSRDPEVVAAYKADPMVHHGKLPAGIARALIGLGQSMPQ RAAALTAPLLVVHGDKDRLIPVAGSRLLVDRVASEDVHLKVYPGLYHEVFNEPEQKLVLDDVTSWIVSHL
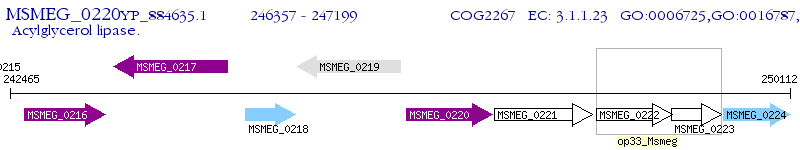
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0220 | - | - | 100% (280) | monoglyceride lipase |
| M. smegmatis MC2 155 | MSMEG_6029 | - | 6e-22 | 27.94% (272) | hypothetical protein MSMEG_6029 |
| M. smegmatis MC2 155 | MSMEG_1576 | - | 2e-13 | 28.78% (271) | alpha/beta hydrolase fold |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0189 | - | 1e-109 | 67.03% (279) | lysophospholipase |
| M. gilvum PYR-GCK | Mflv_0518 | - | 1e-120 | 73.55% (276) | acylglycerol lipase |
| M. tuberculosis H37Rv | Rv0183 | - | 1e-109 | 67.03% (279) | lysophospholipase |
| M. leprae Br4923 | MLBr_02603 | - | 1e-100 | 64.13% (276) | hypothetical protein MLBr_02603 |
| M. abscessus ATCC 19977 | MAB_4551c | - | 7e-95 | 60.81% (273) | lysophospholipase |
| M. marinum M | MMAR_0427 | - | 1e-108 | 68.10% (279) | lysophospholipase |
| M. avium 104 | MAV_4997 | - | 1e-112 | 70.55% (275) | lysophospholipase |
| M. thermoresistible (build 8) | TH_3727 | - | 2e-11 | 26.54% (260) | PUTATIVE Chloride peroxidase |
| M. ulcerans Agy99 | MUL_1077 | - | 1e-107 | 67.38% (279) | lysophospholipase |
| M. vanbaalenii PYR-1 | Mvan_0150 | - | 1e-122 | 74.64% (276) | alpha/beta hydrolase fold |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0518|M.gilvum_PYR-GCK ---MTHSERSFDGLGGVRIVYDVWTPESDSRGVVVLAHGYAEHARRYDHV
Mvan_0150|M.vanbaalenii_PYR-1 -MAVTRTERSFDGVGGVRIVYDVWTPETPPRGIVVLAHGYAEHARRYDHV
MSMEG_0220|M.smegmatis_MC2_155 MVSSTRSEHSFAGVGGVRIVYDVWTPDTDPRGVVVLAHGYAEHAGRYHHV
Mb0189|M.bovis_AF2122/97 -MTTTRTERNFAGIGDVRIVYDVWTPDTAPQAVVVLAHGLGEHARRYDHV
Rv0183|M.tuberculosis_H37Rv -MTTTRTERNFAGIGDVRIVYDVWTPDTAPQAVVVLAHGLGEHARRYDHV
MMAR_0427|M.marinum_M -MNTTRTERTFDGVGGVHIVYDVWTPDAAPKAVVVLAHGLGEHARRYDHV
MUL_1077|M.ulcerans_Agy99 -MNTTRTERTFDGVGGVHIVYDVWTPDAAPKAVVVLAHGLGEHARRYDHV
MAV_4997|M.avium_104 -MN--RTERSFDGVGGVRIVYDVWTPEVAPRAVLVLAHGFGEHARRYDHV
MLBr_02603|M.leprae_Br4923 -MTITRTERNFYGVGGVRIVYDVWMPDTRPRAVIILAHGFGEHARRYDHV
MAB_4551c|M.abscessus_ATCC_199 ------MQRTFDGADDVRIVYDTWTPAGTPRAVVVLSHGFGEHARRYDHV
TH_3727|M.thermoresistible__bu -------MATITTSDGVEIFYRDWGSGQP----IVFSHGWPLTADDWDPQ
.: ..*.*.* * . ::::** * :.
Mflv_0518|M.gilvum_PYR-GCK AARFAESGLITYALDHRGHGRSGGKRVYLRDITEYTGDFHTLVGIARNAY
Mvan_0150|M.vanbaalenii_PYR-1 AARFAEAGLGIYALDHRGHGRSGGKRVYVRDISEYTGDFHSLVRIAAGEH
MSMEG_0220|M.smegmatis_MC2_155 AQRFGAAGLLVYALDHRGHGRSGGKRVHLRDLSEFVEDFRTLVGIAANDH
Mb0189|M.bovis_AF2122/97 AQRLGAAGLVTYALDHRGHGRSGGKRVLVRDISEYTADFDTLVGIATREY
Rv0183|M.tuberculosis_H37Rv AQRLGAAGLVTYALDHRGHGRSGGKRVLVRDISEYTADFDTLVGIATREY
MMAR_0427|M.marinum_M AQRLGAAGLVTYALDHRGHGRSGGKRVLVRDISEYTADFDTLVGIATRDN
MUL_1077|M.ulcerans_Agy99 AQRLGAAGLVTYTLDHRGHGRSGGKRVLVRDISEYTADFDTLVGIATRDN
MAV_4997|M.avium_104 ARRFGAAGLVTYALDHRGHGRSGGKRVLVRDIHEYTTDFDTLVGIATREH
MLBr_02603|M.leprae_Br4923 AHYFAAAGLATYALDLRGHGRSAGKRVLVRDLSEYNADFDILVGIATRDH
MAB_4551c|M.abscessus_ATCC_199 ARRFNEAGYLVYALDHRGHGRSGGKRVYLRDISEYTDDFGTLVDIAAREH
TH_3727|M.thermoresistible__bu MLFFLNAGYRVIAHDRRGHGRSS-PVPDGHDMDHYADDLAALVEHLGLR-
: :* : * ******. :*: .: *: **
Mflv_0518|M.gilvum_PYR-GCK PHLKLIVLGHSMGGGVVFTYGVEHPDD---YDAMVLSGPAVNAHDSVPA-
Mvan_0150|M.vanbaalenii_PYR-1 PGRKLVVLGHSMGGGVVFTYGVEHPDD---YDAMVLSGPAVDAHSSVSP-
MSMEG_0220|M.smegmatis_MC2_155 PTLPRIVLGHSMGGGIVFAYGARYPGE---YSAMVLSGPAVNAHDGVSP-
Mb0189|M.bovis_AF2122/97 PGCKRIVLGHSMGGGIVFAYGVERPDN---YDLMVLSAPAVAAQDLVSP-
Rv0183|M.tuberculosis_H37Rv PGCKRIVLGHSMGGGIVFAYGVERPDN---YDLMVLSAPAVAAQDLVSP-
MMAR_0427|M.marinum_M PGLKCIVLGHSMGGGIVFAYGVERPDN---YDLMVLSAPAVAAQDLVSP-
MUL_1077|M.ulcerans_Agy99 PGLKCIVLGHSMGGGIVFAYGVERPDN---YDLMVLSAPAVAAQDLVSP-
MAV_4997|M.avium_104 HGLKCIVVGHSMGGGIVFAYGVERPDN---YDLMVLSGPAVAAQDQVSP-
MLBr_02603|M.leprae_Br4923 PGLKRIVAGHSMGGAIVFAYGVERPDN---YDLMVLSGPAVAAQDMVSP-
MAB_4551c|M.abscessus_ATCC_199 PDLKRIVLGHSMGGGIVFAYGVDHQDR---YDLMVLSGPAIAAQVGLPY-
TH_3727|M.thermoresistible__bu ---DAVHVGHSTGGGVVVRYLARHGEERTAKAVLIAAVPPLMVRTDANPG
: *** **.:*. * . :: : *.: .:
Mflv_0518|M.gilvum_PYR-GCK ------------VKLVMAKVLGRIAPGLPVENLPADAVSRDPQVVSDYEN
Mvan_0150|M.vanbaalenii_PYR-1 ------------VMVLLAKVLGRLSPGLPVENLPADAVSRDPQVVAAYEN
MSMEG_0220|M.smegmatis_MC2_155 ------------VLVAVAKVLGKLAPGIPVENLDADAVSRDPEVVAAYKA
Mb0189|M.bovis_AF2122/97 ------------VVAVAAKLLGVVVPGLPVQELDFTAISRDPEVVQAYNT
Rv0183|M.tuberculosis_H37Rv ------------VVAVAAKLLGVVVPGLPVQELDFTAISRDPEVVQAYNT
MMAR_0427|M.marinum_M ------------VIAAAAKVLGVVVPGLPVQELDFTAISRDPEVVAAYQN
MUL_1077|M.ulcerans_Agy99 ------------VIAAAAKVLGVVVPGLPVQELDFTAISRDPEVVAAYQN
MAV_4997|M.avium_104 ------------LLALAAKVLGVIVPGLPAQELDADAVSRDPEVVAAYRN
MLBr_02603|M.leprae_Br4923 ------------LRAVVGKGLGLVAPGLPVHQLEVDAISRNRAVVAAYKD
MAB_4551c|M.abscessus_ATCC_199 ------------VLTLVAPVVGRLAPGLPVQKLDVNAISHDPAIIAAYHA
TH_3727|M.thermoresistible__bu GLPKSVFDDFQAQLAANRSEFYRALASGPFYGFNRDGVEPSEALIANWWR
. .. * : .:. . :: :
Mflv_0518|M.gilvum_PYR-GCK DPLVHHGKLPAGVGRALIAVGETMPARAAAITAPLLVVHGDKDRLIPVAG
Mvan_0150|M.vanbaalenii_PYR-1 DPLVHHGKLPAGVGRALIGVGETMPARAAAITAPLLIVHGDNDKLIPVEG
MSMEG_0220|M.smegmatis_MC2_155 DPMVHHGKLPAGIARALIGLGQSMPQRAAALTAPLLVVHGDKDRLIPVAG
Mb0189|M.bovis_AF2122/97 DPLVHHGRVPAGIGRALLQVGETMPRRAPALTAPLLVLHGTDDRLIPIEG
Rv0183|M.tuberculosis_H37Rv DPLVHHGRVPAGIGRALLQVGETMPRRAPALTAPLLVLHGTDDRLIPIEG
MMAR_0427|M.marinum_M DPQVYHGRVPAGIGRALLQVGETMPRRAPALTAPLLVIHGTDDRLIPIEG
MUL_1077|M.ulcerans_Agy99 DPQVYHGRVPAGIGRALLQVGETMPRRAPALTAPLLVIHGTDDRLIPIEG
MAV_4997|M.avium_104 DPLVYHGKVPAGVGRALLQVGETMPQRAPALTAPLLVVHGSDDRLIPVAG
MLBr_02603|M.leprae_Br4923 DPLVYHGKVPAGVGRVMLQVGETMTRRAPVLTTPLLVVHGSEDRLVLVDG
MAB_4551c|M.abscessus_ATCC_199 DPLVHHGRVPAGIGRALLGVGKTMRQRAAGLKRPVLTMHGSDDRLTAPEG
TH_3727|M.thermoresistible__bu QG--MMGDAKAHYDGIVAFSQTDFTEDLTKITVPVLVMHGDDDQVVPYAN
: * * : : . :. *:* :** .*:: .
Mflv_0518|M.gilvum_PYR-GCK SRQLMECIGSPDAHLKVYPGLYHEVFNEPEKELVLDDVTSWIESKL
Mvan_0150|M.vanbaalenii_PYR-1 SRKLVDRVGSADVHLKEYPGLYHEVFNEPEKALVLDDVTSWIESQL
MSMEG_0220|M.smegmatis_MC2_155 SRLLVDRVASEDVHLKVYPGLYHEVFNEPEQKLVLDDVTSWIVSHL
Mb0189|M.bovis_AF2122/97 SRRLVECVGSADVQLKEYPGLYHEVFNEPERNQVLDDVVAWLTERL
Rv0183|M.tuberculosis_H37Rv SRRLVECVGSADVQLKEYPGLYHEVFNEPERNQVLDDVVAWLTERL
MMAR_0427|M.marinum_M SRRLVGHVGSADVELKEYPGLYHEAFNEPERDQVLDDVVSWITARL
MUL_1077|M.ulcerans_Agy99 SRRLVGHVGSADVELKEYPGPYHEAFNEPERDQVLDDVVSWITARL
MAV_4997|M.avium_104 SRRLVECVGSADVELKVYPGLYHEVFNEPEREQVLDDVVGWITARL
MLBr_02603|M.leprae_Br4923 SHRLVECVGSTDVELKVYPGLYHEVFNEPERDQVLEDVVCWILKRL
MAB_4551c|M.abscessus_ATCC_199 SRWLSESAP--DATLKIWNGLYHEIFNEFEKELVLDEVVGWIDARL
TH_3727|M.thermoresistible__bu SGPLTAELLR-NAKLKTYQGFPHGMMTT-HAEVVNADILEFLRS--
* * :. ** : * * :. . * :: ::