For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSSVVVVGSGVVGLCTAWNLARAGAEVTVVSGLDPEREIGWGSGAWVNASSKVRLGYPDHYTLLNQRGMT ALHHLAEKLTDERWLHITGAVEIVSGDDQRRTLAEDVRRLVEFDYPAELLDDADAARVMPGVRLADDEAA AVFPAEGWVDIALLLTTLTAAIAEAGGRFVRRTVTGFVDDAGTVTGVGLDDGSVLSADRYVIAAGAWTGQ LAALAGIDIPVLPADHPKVPGLAVAVTSPANESFPILVTPEVVARPSGPNRMLLAGDSHGIALTIDSPRS DLIRAAEVLLARATARVPALAASAILDVRLSLRAIPSDGITIAGFPAGRDDVYVLTTHSGFSLAAVLGDL AAREILTGQEQQALHEYRPTRFSANASA
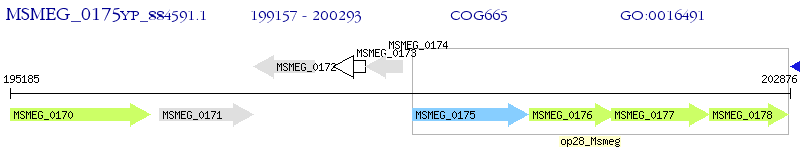
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0175 | - | - | 100% (378) | FAD dependent oxidoreductase, putative |
| M. smegmatis MC2 155 | MSMEG_6264 | - | 5e-15 | 27.53% (396) | putative oxidoreductase |
| M. smegmatis MC2 155 | MSMEG_6291 | - | 2e-14 | 27.10% (428) | D-amino-acid dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1296 | - | 8e-14 | 26.57% (399) | FAD dependent oxidoreductase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_4420 | gcvT_2 | 6e-07 | 27.06% (255) | aminomethyltransferase GcvT_2 |
| M. avium 104 | MAV_2749 | - | 8e-05 | 25.17% (298) | FAD dependent oxidoreductase domain-containing protein |
| M. thermoresistible (build 8) | TH_2138 | - | 2e-17 | 25.83% (422) | D-amino-acid dehydrogenase |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_2290 | - | 1e-12 | 26.69% (251) | FAD dependent oxidoreductase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4420|M.marinum_M -----------------------MGPKIVIVGAGIVGASLADELTARGVI
Mvan_2290|M.vanbaalenii_PYR-1 ---------------MTSPKALPARAQAVIIGGGVIGTSVAYHLTKLGVT
Mflv_1296|M.gilvum_PYR-GCK ---------------------MTETADVVIVGGGLEGAAAAWALSRRGVT
MSMEG_0175|M.smegmatis_MC2_155 ------------------------MSSVVVVGSGVVGLCTAWNLARAGAE
TH_2138|M.thermoresistible__bu ------------MSQRFAERVDGGPRSAIVIGAGIVGLSTAWFLQERGVD
MAV_2749|M.avium_104 MTSLWMSERTERPWAASQLDQGPASADVIVVGAGITGLMTAALLARAGKD
. :::*.*: * * * *
MMAR_4420|M.marinum_M DVTVVDRGP---LFTTGGSTSHA-------------PGLVFATNPSKTMS
Mvan_2290|M.vanbaalenii_PYR-1 DVVLVEQGQ---LSS--GTTWHA-------------AGLVGQLRASEGMT
Mflv_1296|M.gilvum_PYR-GCK DVVVCERST---VGS--GMTGKS-------------SGIVRCHYGVSSLA
MSMEG_0175|M.smegmatis_MC2_155 VTVVSGLDPEREIGWGSGAWVNA-------------SSKVRLGYP-DHYT
TH_2138|M.thermoresistible__bu VCVVDRTGVAAGASWGNAGWISPGLAIPLNEPGVLRYGLRNLLSPTAPLH
MAV_2749|M.avium_104 VLVLEARTV--------GACATAN------------TTAKISLLQGSQLS
.: . .
MMAR_4420|M.marinum_M AFARYTVDKFSGLHHRDG--AVFNRVGGLEVAT-------TAERWADLHR
Mvan_2290|M.vanbaalenii_PYR-1 RLVQYSTALYSELENETGLSAGYKQCGGVTVAR-------TEDRMVQLRR
Mflv_1296|M.gilvum_PYR-GCK AMAADGLEVFEQAQDIFGTDIGFRQTG-YVVGV-------GEQNVDALRK
MSMEG_0175|M.smegmatis_MC2_155 LLNQRGMTALHHLAEKLTDERWLHITGAVEIVS-------GDDQRRTLAE
TH_2138|M.thermoresistible__bu LPLTRDLRQWAFLTRFAANCRRSSWTRALLANKPLNDEAIEAYDVLTANG
MAV_2749|M.avium_104 KVLPRHGREVTRAYVDGNREGQEWVLGHCEAHG-------VAVQREDAYT
MMAR_4420|M.marinum_M KAGWAQAWGVPGRLVGPDQCVELHPLLDRAGILGGFHTPSDGLASAVRAV
Mvan_2290|M.vanbaalenii_PYR-1 TAANAEAFDLQCELLTPAQALDHYPVMRVDDLVGAIWLPADGKANPTDLT
Mflv_1296|M.gilvum_PYR-GCK SLAAQRSVGVETEEIDESEVARLWPFADLTPFAAFGWEARGGYGDAYQTA
MSMEG_0175|M.smegmatis_MC2_155 DVRRLVEFDYPAELLDDADAARVMPGVRLADDEAAAVFPAEGWVDIALLL
TH_2138|M.thermoresistible__bu VDAPVTDAPITAVFRDPREAGRLRDELRRVAETGRDVEVTELSGADLRER
MAV_2749|M.avium_104 YAQSVRGVPSARAEFEACQAVGLPVVWQDDAEVPFAYHGGVRLADQAQFD
:
MMAR_4420|M.marinum_M EAQARRAIARGAVFLAETEVQRVADHNGLVTGVHTSRG-VIEADIVVCAA
Mvan_2290|M.vanbaalenii_PYR-1 FALAKGARTRGCRIVEKTRVLDVLLADGAVTGVRTGAG-DIEAEIVVNCA
Mflv_1296|M.gilvum_PYR-GCK QAFSTSARAAGVRVRQGTAVAGLLADADRVTGVRLADGSEISAGTTVVAT
MSMEG_0175|M.smegmatis_MC2_155 TTLTAAIAEAGGRFVRR-TVTGFVDDAGTVTGVGLDDGSVLSADRYVIAA
TH_2138|M.thermoresistible__bu VPLASAEITVGLEIHGQRYVDPARFVQALADSVVDRGATIHRMDITGVDS
MAV_2749|M.avium_104 PMPFLDSLAVELLGRGGRLVEHTRVRRVSWRGKGVRVHANQGSDAAGHDV
* .
MMAR_4420|M.marinum_M GFWGAQLAKQVGLVLPLVPMAHQYAKTGQVAPLVGRNTEDAPASLPILRH
Mvan_2290|M.vanbaalenii_PYR-1 GQWAKQVGAMAGVNVPLHSAEHFYVVS--------EKIDGVHPDLPILRD
Mflv_1296|M.gilvum_PYR-GCK GVWTRPFLSAHGIDMPIRVVREQIVMIDP------GIDEAAVGRLPVFSD
MSMEG_0175|M.smegmatis_MC2_155 GAWTGQLAALAGIDIPVLPADHPKVPG-----LAVAVTSPANESFPILVT
TH_2138|M.thermoresistible__bu GPGGVTVHSAGGQELSAGAVVIATGAWLHQLAQRWVRVPVQAGRGYSFTV
MAV_2749|M.avium_104 ELHADQLVLATGIPILDRGGYFARVKPSRSYCLAFKVPGNITRPMMISTD
. * :
MMAR_4420|M.marinum_M QDGDLYFREHVDRLGIGSYGHQPMPVDMSTLLADTAGEPMPSMLPFTEED
Mvan_2290|M.vanbaalenii_PYR-1 PDGYTYFKEEVGGLVIGGFEPEAKPWVSPDAIPYPF---EFQLLDEDWEH
Mflv_1296|M.gilvum_PYR-GCK LVSLQYVRPELGGEILFGNSDLPGYDAIAPADPDAY------LNRATDDF
MSMEG_0175|M.smegmatis_MC2_155 P--EVVARPSGPNRMLLAGDSHGIALTIDSPRSDLIR------------A
TH_2138|M.thermoresistible__bu PVDRPVPAPVYLPEVRVACTPCAAGLRVAGTMEFRDP-----DAPLISKR
MAV_2749|M.avium_104 SPTRSVRYAPVPDGERLIVGGAGHTVGREKSP---------------SAA
MMAR_4420|M.marinum_M FEPAWRECVRLLPALGDTKVDEGFNGIFSFTPDGYSLLGEHR-ELGGFWV
Mvan_2290|M.vanbaalenii_PYR-1 FEILMNSALLRIPALETTGIKKFYNGPESFTPDNQFILGEAP-ECRNFFV
Mflv_1296|M.gilvum_PYR-GCK VDLTVDKVGTRFPGFPDASITGSYAGCYDVTPDWNPVISRG--PLDGLVI
MSMEG_0175|M.smegmatis_MC2_155 AEVLLARATARVPALAASAILDVRLSLRAIPSDGITIAGFPA-GRDDVYV
TH_2138|M.thermoresistible__bu VDAIVAAARPLLDGVRWDERGPSWVGPRPVTPDGRPLIGAV---AEDVYV
MAV_2749|M.avium_104 LDELSAWTRKHFPGAVQT---HFWSAQDYTPIDHLPYVGPILPNSETIFV
: . . . . * . . :
MMAR_4420|M.marinum_M AE---AVWVTHSAGVAKAVAEWIVDG--TPSIDVHECDLYRFEDFARSPR
Mvan_2290|M.vanbaalenii_PYR-1 GAGFNSVGIATAGGAGRALAEWIVNG--APTTDLTGVDIRRFAPFNGNNR
Mflv_1296|M.gilvum_PYR-GCK AAGFSGHGFKIAPAVGRLVADLVVDG--HGSDPRIPASDFRFSRFAEND-
MSMEG_0175|M.smegmatis_MC2_155 LT--THSGFSLAAVLGDLAAREILTG--QEQQALHEYRPTRFSANASA--
TH_2138|M.thermoresistible__bu AAGHGMWGLTHGPVTGRLLAEQITTG--KQPDVLDEFDPLR---------
MAV_2749|M.avium_104 ATGFNKWGMTNGAAAALALSSRILGGRMDWARAFASWSPHELSGLATAVQ
. . . : : * .
MMAR_4420|M.marinum_M FIDETCAQAFVEVYDIVHPHQYRSALRGLRTSPFHHRQRELGAYFYEGGG
Mvan_2290|M.vanbaalenii_PYR-1 WLHDRVAEVLGIHYEIPWPNREMKTARPFRRSPVHHLLDAAGANFGSRMG
Mflv_1296|M.gilvum_PYR-GCK ------------PLKTPYPYVGAGQMR-----------------------
MSMEG_0175|M.smegmatis_MC2_155 --------------------------------------------------
TH_2138|M.thermoresistible__bu --------------------------------------------------
MAV_2749|M.avium_104 ANLEVGFNLTKGWITPAVRIGRRSPVDGDGGVVSGPPWHLQARCRVDGTE
MMAR_4420|M.marinum_M WERPAWFQANAELVRELAGRNVAFPERDDWAARYWSPIAIAEAQWTRERV
Mvan_2290|M.vanbaalenii_PYR-1 WERANFFAPPGEEP----------VIDYSWGKPNWLDWSAAEQASTRTGV
Mflv_1296|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_0175|M.smegmatis_MC2_155 --------------------------------------------------
TH_2138|M.thermoresistible__bu --------------------------------------------------
MAV_2749|M.avium_104 HRVSPVCPHLGG--------------------------------------
MMAR_4420|M.marinum_M ALYDMTPLTRYEVSGAGAVDLLQRLTTNDIDKGVGSVTYTLMLDETGGIR
Mvan_2290|M.vanbaalenii_PYR-1 TVFDQTSFSKYLLVGGDAEAALQWLCTADVGVEVGRSVYTGMLNARGTYE
Mflv_1296|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_0175|M.smegmatis_MC2_155 --------------------------------------------------
TH_2138|M.thermoresistible__bu --------------------------------------------------
MAV_2749|M.avium_104 -----------------------IVNWNDADKAWECPLHGSRFAPDGTLL
MMAR_4420|M.marinum_M SDLTVARLGTARFQVGANGPQD-------FDWLSRHLPQDGSVTLRDITG
Mvan_2290|M.vanbaalenii_PYR-1 SDVTVTRTGQDEYLIVSSAATTERDKDHIRSQIRRRWPDGANAHLVDVTS
Mflv_1296|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_0175|M.smegmatis_MC2_155 --------------------------------------------------
TH_2138|M.thermoresistible__bu --------------------------------------------------
MAV_2749|M.avium_104 EGPATRDLTASR--------------------------------------
MMAR_4420|M.marinum_M ATCCIGVWGPAARDLVQPLCRDDLSHQAFPYFRLLQTYLEAIPVTMLRVS
Mvan_2290|M.vanbaalenii_PYR-1 AYAVFGVMGPRSREVLSALTDADLSDAAFPFGTSRQISLGYATVRATRIT
Mflv_1296|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_0175|M.smegmatis_MC2_155 --------------------------------------------------
TH_2138|M.thermoresistible__bu --------------------------------------------------
MAV_2749|M.avium_104 --------------------------------------------------
MMAR_4420|M.marinum_M YVGELGWEIYTEASYGGALWDLLWAAGAGHQAIAAGRIAFNSLRMEKGYR
Mvan_2290|M.vanbaalenii_PYR-1 YVGELGWELYVPAEFAVGVYEDLLAAGQRHGVARGGYYAIESLRLEKGYR
Mflv_1296|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_0175|M.smegmatis_MC2_155 --------------------------------------------------
TH_2138|M.thermoresistible__bu --------------------------------------------------
MAV_2749|M.avium_104 --------------------------------------------------
MMAR_4420|M.marinum_M AWGTDMTAEHRPDEAGLGFAVRMS--KDFIGKAALVDAAAR---KVLQCL
Mvan_2290|M.vanbaalenii_PYR-1 AFGRELTPSENPVEAGLLFACKLKTDIAFLGRDAVERARTEGPRRRLVSF
Mflv_1296|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_0175|M.smegmatis_MC2_155 --------------------------------------------------
TH_2138|M.thermoresistible__bu --------------------------------------------------
MAV_2749|M.avium_104 --------------------------------------------------
MMAR_4420|M.marinum_M VFDDPAAAALGKEPVLAGDVCIGFVTSAGYSPTIGRTIAYAWLPAES---
Mvan_2290|M.vanbaalenii_PYR-1 RVDSPEPMLWGGELILRDGAVAGQATSAAWGQTVGAAVGLAYLRSGDGAV
Mflv_1296|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_0175|M.smegmatis_MC2_155 --------------------------------------------------
TH_2138|M.thermoresistible__bu --------------------------------------------------
MAV_2749|M.avium_104 --------------------------------------------------
MMAR_4420|M.marinum_M -----SIGDTVTVDHRGRRYPASVHQQPVVDPESTRIRR
Mvan_2290|M.vanbaalenii_PYR-1 VDPDWVRSGSYQINVGGALYPITVSLRPLYDPTGDRVR-
Mflv_1296|M.gilvum_PYR-GCK ---------------------------------------
MSMEG_0175|M.smegmatis_MC2_155 ---------------------------------------
TH_2138|M.thermoresistible__bu ---------------------------------------
MAV_2749|M.avium_104 ---------------------------------------