For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAIASPVSERLTVTALLLLTVATGLVDAISVLVLGHVFVANMTGNVIFLGFWFVPHSGVDLTAAVVAFPA FVVGTIIGGRLARHLDGQVRTWLTTALGVEVVLLTVLSILAGTGVLDYQDNTKLFLIAGLAITFGIQNAT ARQFGIQELSTTVLTSTIVGLGFDSRLAGGTGEREKLRYAVVATMCLGAVIGATLSRFTVAPVIALAAVV IATSACIFRFGR
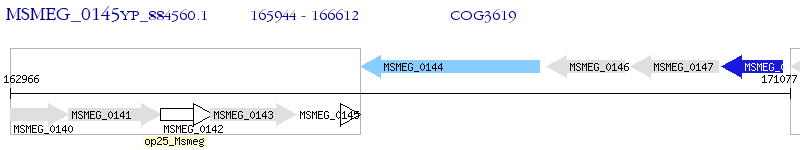
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0145 | - | - | 100% (222) | hypothetical protein MSMEG_0145 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0133 | - | 5e-07 | 28.50% (207) | transmembrane protein |
| M. gilvum PYR-GCK | Mflv_0701 | - | 5e-22 | 65.33% (75) | hypothetical protein Mflv_0701 |
| M. tuberculosis H37Rv | Rv0128 | - | 5e-07 | 28.50% (207) | transmembrane protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3210c | - | 4e-06 | 26.03% (219) | hypothetical protein MAB_3210c |
| M. marinum M | MMAR_2141 | - | 5e-08 | 27.01% (211) | hypothetical protein MMAR_2141 |
| M. avium 104 | MAV_5004 | - | 2e-78 | 68.42% (209) | hypothetical protein MAV_5004 |
| M. thermoresistible (build 8) | TH_2076 | - | 2e-88 | 75.57% (221) | conserved hypothetical protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_1320 | - | 8e-21 | 30.62% (209) | hypothetical protein Mvan_1320 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0133|M.bovis_AF2122/97 ------------------------------------MQREIYDG-EARLS
Rv0128|M.tuberculosis_H37Rv ------------------------------------MQREIYDG-EARLS
MMAR_2141|M.marinum_M ---------------------------------MSWVMREMFDR-EARLS
MAB_3210c|M.abscessus_ATCC_199 MNRKGRSATLESGPQGEIEIGSDKGAIAPLKGDDAVITREAAIGSEARLS
MSMEG_0145|M.smegmatis_MC2_155 ----------------------------------MAIASPVSERLTVTAL
TH_2076|M.thermoresistible__bu ----------------------------------MAVASPVTPRLTVLAL
MAV_5004|M.avium_104 ----------------------------------------------MAAL
Mvan_1320|M.vanbaalenii_PYR-1 ------------------------------------MMADAPADRALAAT
Mflv_0701|M.gilvum_PYR-GCK --------------------------------------------------
Mb0133|M.bovis_AF2122/97 WVLAALAGILGATAFTHSAGYFVTFMTGNSQRAVLGLF--GDDAWMS---
Rv0128|M.tuberculosis_H37Rv WVLAALAGILGATAFTHSAGYFVTFMTGNSQRAVLGLF--GDDAWMS---
MMAR_2141|M.marinum_M WILAALAGLVGAAAFTHTAGYFVTFMTGNSERAVLGFF--TNDRWLS---
MAB_3210c|M.abscessus_ATCC_199 WLLSGLAGMVGAVAFLHSAGYFVTFMTGNTERAVLTWFKVTDKQQVAGAG
MSMEG_0145|M.smegmatis_MC2_155 LLLTVATGLVDAISVLVLGHVFVANMTGN--VIFLGFWFVPHSGVDL---
TH_2076|M.thermoresistible__bu LLMTFATGLVDAVSVLVLGHVFVANMTGN--VIFLGFWFVPTSGVDL---
MAV_5004|M.avium_104 LLLTFATGLADAISILVLGHVFVANMTGN--VIFLGFWLAPRTSIDL---
Mvan_1320|M.vanbaalenii_PYR-1 VALTFVTGVVDAVGFLGLDRVFTGNMTGN--IVILGMGVAGADELPV---
Mflv_0701|M.gilvum_PYR-GCK --MRSACWCWDMCSSRTLPERCVPRLFGT----------CHIPGVDL---
: . . . : *.
Mb0133|M.bovis_AF2122/97 -VTASLLILFFVAGVVIASVCRRHFWAAHP----HGPTVLTTFSLIFAAG
Rv0128|M.tuberculosis_H37Rv -VTASLLILFFVAGVVIASVCRRHFWAAHP----HGPTVLTTFSLIFAAG
MMAR_2141|M.marinum_M -ISAALLLASFVAGVVIASLCRQHIWVDHP----HGPTVLTTLCLGIAAA
MAB_3210c|M.abscessus_ATCC_199 PIAAISMIGAFLFGVIVASYCRRKFWKNHP----HGATVLTTVGLLAAAA
MSMEG_0145|M.smegmatis_MC2_155 -TAAVVAFPAFVVGTIIGGRLARHLDGQVR----TWLTTALGVEVVLLTV
TH_2076|M.thermoresistible__bu -TAAVVAFLAFLVGTVIGGRLARHLAHHVR----GWITSALCVETVLLLA
MAV_5004|M.avium_104 -TAVVVALPTFACTTILSGRLARHFGERVR----AWISTVLATEIVLLVG
Mvan_1320|M.vanbaalenii_PYR-1 -LGPAVALGAFTAAAYLAGLVLRGPGGARQPGWQHRVTVLLTLGAATLTV
Mflv_0701|M.gilvum_PYR-GCK -TAALVAFVGFFDGTIIGGRLSRHLDSNVR----VWLTTALGLEVVALAV
: * . :.. : :
Mb0133|M.bovis_AF2122/97 VDIML-GGWHES--MLDFVPILFVVFGIGALNTSFVKDGEVSVPLSYVTG
Rv0128|M.tuberculosis_H37Rv VDIML-GGWHES--MLDFVPILFVVFGIGALNTSFVKDGEVSVPLSYVTG
MMAR_2141|M.marinum_M LDIVV-HGWSTG--RVTFLPILAVTFGIGALNTSFVKNGEVSISLSYMTG
MAB_3210c|M.abscessus_ATCC_199 VDFYQ-DRSFTANIDVHFTPILLLAFSIGALNTSFVKKGETAIPLSYVTG
MSMEG_0145|M.smegmatis_MC2_155 LSILAGTGVLDYQDNTKLFLIAGLAITFGIQNATARQFGIQELSTTVLTS
TH_2076|M.thermoresistible__bu LSALAGAGWLDYHNDQKFILIAGLAVAFGLQNASARRFGIQELSTTVLTS
MAV_5004|M.avium_104 LSVLAGSGILSYQQDSKLLMIGILAVTFGLQHSSARQFGIQELSTTVLTS
Mvan_1320|M.vanbaalenii_PYR-1 LTVVAVVTGRHPEPVVQIPMAAATAAVMGSQAMVARAVAVADMTTVVVTS
Mflv_0701|M.gilvum_PYR-GCK LAAVAGANLVHYHDYSKLILIVGLAVVSAARTQPPGSLASRCAPRFCRPY
: : . . . . .
Mb0133|M.bovis_AF2122/97 TLVKMGQGIERHLAG--GKVEDWLGYFLLHASFVLGAAAGGAISMVVTGP
Rv0128|M.tuberculosis_H37Rv TLVKMGQGIERHLAG--GKVEDWLGYFLLHASFVLGAAAGGAISMVVTGP
MMAR_2141|M.marinum_M TLVKLGQGIQRHLSG--GQATAWLGYLLLYASYIVGATVGGVLSLVIGGS
MAB_3210c|M.abscessus_ATCC_199 TVVKLGQGIERHLFGR-GSVFEWLGYAMLYLSFVLGAATGGVIASVYSGS
MSMEG_0145|M.smegmatis_MC2_155 TIVGLG--FDSRLAG--GTGEREKLRYAVVATMCLGAVIGATLSRFTVAP
TH_2076|M.thermoresistible__bu TIVGIG--FDSRLAG--GTGKREKLRYTVVLTMLGGAMVGATMTLVTVAP
MAV_5004|M.avium_104 TIVSLG--LDSRLAG--GTGERQRLRIGVVATMCAGAFLGATMSRYVVAP
Mvan_1320|M.vanbaalenii_PYR-1 TLASLAG--ETWTRG--GRGAVANRRLAAIVVIFAGAVVGALLMRIHIAV
Mflv_0701|M.gilvum_PYR-GCK SPESLRPLFRAGGCASHSLASPDNHDQGNESRAEIAGTDGQWVRQQKVSP
: : . . .. * : .
Mb0133|M.bovis_AF2122/97 QMLAVAAVVCAATTGYTYLHADRRGLVNQKRPQPGKRLFRALRRGELDSG
Rv0128|M.tuberculosis_H37Rv QMLAVAAVVCAATTGYTYLHADRRGLVNQKRPQPGKRLFRALRRGELDSG
MMAR_2141|M.marinum_M QMLAVATVACALTTGYTYFHADRRALLKERNGE--RRSGKVIADNDLERV
MAB_3210c|M.abscessus_ATCC_199 KVILTVGIICGATTLVTFFYSDRKTILG----------------------
MSMEG_0145|M.smegmatis_MC2_155 VIALAAVVIATSACIFRFGR------------------------------
TH_2076|M.thermoresistible__bu VIALAAAFVAVSAAIFWSGPSPTSENDL----------------------
MAV_5004|M.avium_104 VFVVAAAVIAASLLIFRFGPPAAKPSAAEAN-------------------
Mvan_1320|M.vanbaalenii_PYR-1 PFGLAAATTAAVALLGHLRLSPPVRSR-----------------------
Mflv_0701|M.gilvum_PYR-GCK RSIEDFRSVVESDRIFQL--------------------------------
Mb0133|M.bovis_AF2122/97 TSTPATNYGSS
Rv0128|M.tuberculosis_H37Rv TSTPATNYGSS
MMAR_2141|M.marinum_M VADEAPDRSE-
MAB_3210c|M.abscessus_ATCC_199 -----------
MSMEG_0145|M.smegmatis_MC2_155 -----------
TH_2076|M.thermoresistible__bu -----------
MAV_5004|M.avium_104 -----------
Mvan_1320|M.vanbaalenii_PYR-1 -----------
Mflv_0701|M.gilvum_PYR-GCK -----------