For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MEGDAGASQLNPTDQQQEHTEPQIEQTTVEADARRPSRLGRGWMATVCAALVLVAAALGVGGYLALRSHL DSNVIARDEAEAMQAAKDCVTATQAPDTAAMAAAQAKIIECATGDFGAQAVLYSGILLDAYQAANVQVQV SDMRAAVEKHNDDGSMDLLVAVRVLVTNTEKANEEQGYRLRVKMARDEDGLYKVARLDQVTS
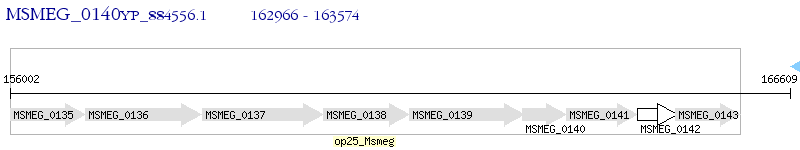
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0140 | - | - | 100% (202) | mce associated membrane protein |
| M. smegmatis MC2 155 | MSMEG_2864 | - | 6e-09 | 31.79% (173) | hypothetical protein MSMEG_2864 |
| M. smegmatis MC2 155 | MSMEG_4771 | - | 3e-05 | 22.50% (200) | hypothetical protein MSMEG_4771 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0181 | - | 2e-58 | 56.46% (209) | mce associated membrane protein |
| M. gilvum PYR-GCK | Mflv_0705 | - | 2e-54 | 52.61% (211) | putative MCE associated membrane protein |
| M. tuberculosis H37Rv | Rv0175 | - | 2e-58 | 56.46% (209) | mce associated membrane protein |
| M. leprae Br4923 | MLBr_02595 | - | 9e-52 | 59.39% (165) | hypothetical protein MLBr_02595 |
| M. abscessus ATCC 19977 | MAB_4147c | - | 4e-05 | 25.64% (195) | MCE-family protein |
| M. marinum M | MMAR_0418 | - | 3e-58 | 56.50% (200) | Mce associated membrane protein |
| M. avium 104 | MAV_5009 | - | 1e-56 | 52.56% (234) | hypothetical protein MAV_5009 |
| M. thermoresistible (build 8) | TH_0291 | - | 6e-54 | 57.29% (192) | PROBABLE CONSERVED MCE ASSOCIATED MEMBRANE PROTEIN |
| M. ulcerans Agy99 | MUL_1068 | - | 4e-58 | 56.00% (200) | Mce associated membrane protein |
| M. vanbaalenii PYR-1 | Mvan_0140 | - | 3e-53 | 50.70% (215) | putative MCE associated membrane protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0181|M.bovis_AF2122/97 ---------MKAADSAESDAGA---DQTG-------------PQVKAADS
Rv0175|M.tuberculosis_H37Rv ---------MKAADSAESDAGA---DQTG-------------PQVKAADS
MMAR_0418|M.marinum_M ---------MTTEDSAESEAAP---DSTDQPEPEAAAEPEASAGSDDAAQ
MUL_1068|M.ulcerans_Agy99 -----MRTEATAEDSSESEAAP---DSTEQPEP------EAAAGSHDAAQ
MLBr_02595|M.leprae_Br4923 --------------MPESQVAA---DDS----------------------
MAV_5009|M.avium_104 MEGDAGASRLNPIDTDDDSLSP---QDKDEDSTAPAS-TDEQTGSQGAPG
MSMEG_0140|M.smegmatis_MC2_155 ----------MEGDAGASQLNP---TDQQ---------------------
Mflv_0705|M.gilvum_PYR-GCK ----------MEGDAGTSRLIP--EADTA-------------DEVVDTP-
Mvan_0140|M.vanbaalenii_PYR-1 ----------MEGDAGTSRLIPDPETDTA-------------DGAQDAPD
TH_0291|M.thermoresistible__bu ----------MEGDAGTRRLNR--TTDAD-------------DAPGESLA
MAB_4147c|M.abscessus_ATCC_199 ---------MTEEKKSASQTTRRRAARPAG----------PTGEVSAEPI
Mb0181|M.bovis_AF2122/97 AESDAGELGEDACPEQALVERRPSRLRRGWLVGIAATLLALAGGLGAAGY
Rv0175|M.tuberculosis_H37Rv AESDAGELGEDACPEQALVERRPSRLRRGWLVGIAATLLALAGGLGAAGY
MMAR_0418|M.marinum_M EQAEDTEQSGDGQAVEPGVERRPSRLGRGWLVGISAVLLLAAGGIGAGGY
MUL_1068|M.ulcerans_Agy99 EQAEDTEQSGDGQAVEPGVERRPSRLGRGWLVGISAVLLLAAGGIGAGGY
MLBr_02595|M.leprae_Br4923 -----------------GVTLNRSRLGRGWLTGIAVALLLAGCGIGTGGY
MAV_5009|M.avium_104 VAGESAPDDADDAVPVAAVPRGPSRLGRGWLAAIAAVLVLCAAGIGAGGY
MSMEG_0140|M.smegmatis_MC2_155 ---QEHTEPQIEQTTVEADARRPSRLGRGWMATVCAALVLVAAALGVGGY
Mflv_0705|M.gilvum_PYR-GCK --TAEASDPSPDQ-DVPADTRRPTRLGNGWVATICAVLLLLAGGAVAAGV
Mvan_0140|M.vanbaalenii_PYR-1 EASLGAPDTPPQE-QPPAEPRRPSRLGTGWVATICVVLLLLGGGGITAGY
TH_0291|M.thermoresistible__bu GDPTDVTEPSTQTPEPPAEERRPSRLSGTWVAGICAALVVGAAGLAAGGY
MAB_4147c|M.abscessus_ATCC_199 SVSLGRTEALSKPEPTVADSRRPGNT-RGVLRAATVLVVVLVGALGVALA
. : . :: . ..
Mb0181|M.bovis_AF2122/97 FALRSHQESQSIAREDLAAIEAAKDCVAATQAPDAGAMSASMQKIIECGT
Rv0175|M.tuberculosis_H37Rv FALRSHQESQSIAREDLAAIEAAKDCVAATQAPDAGAMSASMQKIIECGT
MMAR_0418|M.marinum_M LALRSHQESQVIARNDAAALEAAKDCVAATQAPDTSAMAASEQKIIDCGT
MUL_1068|M.ulcerans_Agy99 LALRSHQESQVIARNDAAALEAAKDCVAATQAPDTSAMAASEQKIIDCGT
MLBr_02595|M.leprae_Br4923 CMLRYHQDSQAMARNDNAALKTALDCVAATQAPDTNTMAASEQKIIDCGT
MAV_5009|M.avium_104 LALRYHHESQVIARNNAAALKAAVDCVEATQAPDTNAMTASEQKIIDCGT
MSMEG_0140|M.smegmatis_MC2_155 LALRSHLDSNVIARDEAEAMQAAKDCVTATQAPDTAAMAAAQAKIIECAT
Mflv_0705|M.gilvum_PYR-GCK LSLRANERAAETARSDEAALQAAKDCVAATQAPDTAAMTAAQSKILECST
Mvan_0140|M.vanbaalenii_PYR-1 LALRANARAAEVARSDDAALQAAKECVAATQAPDTAAMTAAQSKILECST
TH_0291|M.thermoresistible__bu LAWRSHERSVDIARIEAAALEAAKDCVAATHAPDVESMATSQAKIIECST
MAB_4147c|M.abscessus_ATCC_199 LMGRAQHRDDLRAARDQRFVDTAVQTVTNMMTYSPDSIDKNVDQFVNSTS
* : * : :.:* : * : . :: ::::. :
Mb0181|M.bovis_AF2122/97 GDFG-AQASLYTSMLVEAYQAASVHVQVTDMRAAVERNNND-GSVDVLVA
Rv0175|M.tuberculosis_H37Rv GDFG-AQASLYTSMLVEAYQAASVHVQVTDMRAAVERNNND-GSVDVLVA
MMAR_0418|M.marinum_M DQYR-SQAVLYSSMLVQAYQAANVHVQLTDMRAAVERNNPD-GSVDVLVA
MUL_1068|M.ulcerans_Agy99 DQYR-SQAVLYSSMLVQAYQAANVHVQLTEMRAAVERNNPD-GSVDVLVA
MLBr_02595|M.leprae_Br4923 DAFH-AQALLYTNMLVQAYQAANVHVQVSDMRAAVERHNND-GSIDVLVA
MAV_5009|M.avium_104 DAYR-SQALLYTSMLVQAYQSANVHVQVSDVRAAVERNNPD-GSIDILVA
MSMEG_0140|M.smegmatis_MC2_155 GDFG-AQAVLYSGILLDAYQAANVQVQVSDMRAAVEKHNDD-GSMDLLVA
Mflv_0705|M.gilvum_PYR-GCK GEFR-VQAGLFSSIVAEAYQAANATVAVDDLRAAVERHNED-GTVNVLVA
Mvan_0140|M.vanbaalenii_PYR-1 GEFG-VQAGLFSSVLAEAYQAADATVAVDDLRAAVERRNED-GTVNVLVA
TH_0291|M.thermoresistible__bu GDFA-VQANLYSGMLVDAYRVADASVQVSDMRAAVERHHDD-GVVDVLVA
MAB_4147c|M.abscessus_ATCC_199 GPLRGTFNPDNTANLKALYHQTGTSSEVVIKKASLESIDEVSKKASILIS
. : : *: :.. : :*::* . .:*::
Mb0181|M.bovis_AF2122/97 LRVKVSNTDSDAHEVGYRL----------RVRMALDE-GRYKIAKLDQVT
Rv0175|M.tuberculosis_H37Rv LRVKVSNTDSDAHEVGYRL----------RVRMALDE-GRYKIAKLDQVT
MMAR_0418|M.marinum_M LRVKMSNDQDQNQETGYRL----------RVRMTPTE-GQYKISKLDQVA
MUL_1068|M.ulcerans_Agy99 LRVKMSNDQDQNQETGYRL----------RVRMTPTE-GQYKISKLDQVA
MLBr_02595|M.leprae_Br4923 LRVKLSNDRAHNQETGYRL----------RVKMALAE-GQYKISKLDQVT
MAV_5009|M.avium_104 LRVKVASDQSQN-ESGYRL----------RVKMALAE-GQYKISKLDQVT
MSMEG_0140|M.smegmatis_MC2_155 VRVLVTNTEKANEEQGYRL----------RVKMARDEDGLYKVARLDQVT
Mflv_0705|M.gilvum_PYR-GCK VRVRITNAEAADQRVGYRL----------RATMAWDE-AQYRISSLEQVN
Mvan_0140|M.vanbaalenii_PYR-1 VRVRISNSEAADQRVGYRL----------RATMAFDE-GQYRISSLDQVN
TH_0291|M.thermoresistible__bu MRVKVTNTAAADQEHGYRLXXRRRPYLLPRFRRAHDR-SATVRTPKSQIT
MAB_4147c|M.abscessus_ATCC_199 ARVTLTNADSGVNEPSKSY----------RMRMIVAEDQQGNMTAYDLRW
** ::. . . * . : .
Mb0181|M.bovis_AF2122/97 K-----
Rv0175|M.tuberculosis_H37Rv K-----
MMAR_0418|M.marinum_M K-----
MUL_1068|M.ulcerans_Agy99 K-----
MLBr_02595|M.leprae_Br4923 K-----
MAV_5009|M.avium_104 K-----
MSMEG_0140|M.smegmatis_MC2_155 S-----
Mflv_0705|M.gilvum_PYR-GCK S-----
Mvan_0140|M.vanbaalenii_PYR-1 S-----
TH_0291|M.thermoresistible__bu SAGRVP
MAB_4147c|M.abscessus_ATCC_199 PDGQS-