For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAMEERRERERAARRDLIISTARRLAEAEGWQAVTTRRLSAEIEYSQPVLYKHFAGMEELADAVALDGFG ELAEAMRQARRGDDHPDHAVTRIAQAYLDFARANPAVYDAMFARATNLRFAADDTPSPLTEAFAQLRDAV GRNEDADTLTEVLWAALHGLVMLGRTGRLRPDHETERLQLLVDRMSR
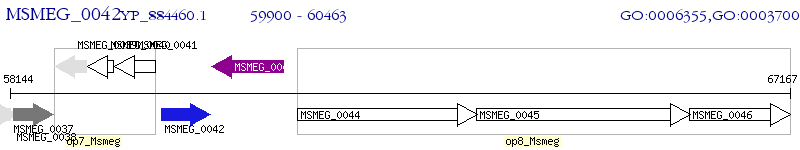
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0042 | - | - | 100% (187) | TetR-family protein transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_1397 | - | 1e-11 | 29.17% (168) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_5177 | - | 1e-07 | 24.70% (166) | regulatory protein, TetR |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0700 | - | 6e-10 | 27.67% (159) | TetR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_2110 | - | 4e-14 | 35.09% (171) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv0681 | - | 6e-10 | 27.67% (159) | TetR family transcriptional regulator |
| M. leprae Br4923 | MLBr_02568 | - | 4e-06 | 24.70% (166) | putative TetR-family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_3852c | - | 2e-13 | 32.18% (174) | TetR family transcriptional regulator |
| M. marinum M | MMAR_1010 | - | 2e-10 | 28.85% (156) | TetR family transcriptional regulator |
| M. avium 104 | MAV_4494 | - | 2e-10 | 28.11% (185) | transcriptional regulator, TetR family protein |
| M. thermoresistible (build 8) | TH_1893 | - | 8e-12 | 29.41% (170) | PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN (POSSIBLY |
| M. ulcerans Agy99 | MUL_0762 | - | 2e-10 | 28.85% (156) | TetR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_1278 | - | 4e-13 | 28.07% (171) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
TH_1893|M.thermoresistible__bu VNTQMSAGPGGAPGART-KSPARAGRLSRDTIVNAALTFLDREGWDALTI
Mvan_1278|M.vanbaalenii_PYR-1 ----MAVRP------------PRTAKLSRDAIVNAALTFLDREGWDALTI
Mb0700|M.bovis_AF2122/97 --------------------MARPAKLSRESIVEGALTFLDREGWDSLTI
Rv0681|M.tuberculosis_H37Rv --------------------MARPAKLSRESIVEGALTFLDREGWDSLTI
MAV_4494|M.avium_104 -MAAQPEPPTAAGRQRAGRPAARPAKLSREGIIDGALTFLDREGWDALTI
MMAR_1010|M.marinum_M -MAASAEGPSAAGRPRAAKSTSRPAKLSRDAIVDGALTFLDREGWDSLTI
MUL_0762|M.ulcerans_Agy99 -MAASAEGPSAAGRPRAAKSTSRPAKLSRDAIVDGALTFLDREGWDSLTI
MAB_3852c|M.abscessus_ATCC_199 ----MTTEPATAHR----RTPARATRLNRDAVVNAALSFLDRAGWDALTI
MSMEG_0042|M.smegmatis_MC2_155 ----MAMEE----------RRERERAARRDLIISTARRLAEAEGWQAVTT
Mflv_2110|M.gilvum_PYR-GCK -----------------------MVSTVRDNLVAAGVRLLEGGGVSALSA
MLBr_02568|M.leprae_Br4923 ---------------MAGGAKRIPRAVREQQMLDAAVQMFSVNGYHKTSM
.: :: . : . * :
TH_1893|M.thermoresistible__bu NALATQLGTKGPSLYNHVQSLDDLRRSVRMRVVDDIIGMLNAAGQGR-TR
Mvan_1278|M.vanbaalenii_PYR-1 NALANQLGTKGPSLYNHVHSLDDLRRTVRMHVVGDIIEMLNTVGKGR-TP
Mb0700|M.bovis_AF2122/97 NALATQLGTKGPSLYNHVDSLEDLRRAVRIRVIDDIITMLNRVGAGR-AR
Rv0681|M.tuberculosis_H37Rv NALATQLGTKGPSLYNHVDSLEDLRRAVRIRVIDDIITMLNRVGAGR-AR
MAV_4494|M.avium_104 NALATQLGTKGPSLYNHVDSLEDLRRAVRIRVIDDIITMLNRVGEGR-AR
MMAR_1010|M.marinum_M NALATQLGTKGPSLYNHVVSLEDLRGAVRVRVIDDIIMMLNRVGEGR-SR
MUL_0762|M.ulcerans_Agy99 NALATQLGTKGPSLYNHVVSLEDLRGAVRVRVIDDIIMMLNRVGEGR-SR
MAB_3852c|M.abscessus_ATCC_199 NALAVELGTKGPSLYNHVDSLDDLRHEVRDRVLAEIVGMLHTVSSGR-SS
MSMEG_0042|M.smegmatis_MC2_155 RRLSAEIEYSQPVLYKHFAGMEELADAVALDGFGELAEAMRQARRGDDHP
Mflv_2110|M.gilvum_PYR-GCK RSVATEAGTSTMALYTHFGGMTGLLDAVAVDVFARFTEALTEVAPTD-DP
MLBr_02568|M.leprae_Br4923 DAIAAEAQISKPMLYLYYGSKEDLFGACLNREMSRFIAMVRADIDFTQSP
:: : . ** : . * . : :
TH_1893|M.thermoresistible__bu DDAVLAMASAYRSYAHHHPGRYSAFTRMPLGGDDPEFSAAARAAAGP---
Mvan_1278|M.vanbaalenii_PYR-1 DDAVMVMASAYRSYAHHHPGRYSAFTRMPLGGDDPEFTEATRAAAAP---
Mb0700|M.bovis_AF2122/97 DDAVLVMAGAYRSYAHHHPGRYSAFTRMPLGGDDPEYTAATRGAAAP---
Rv0681|M.tuberculosis_H37Rv DDAVLVMAGAYRSYAHHHPGRYSAFTRMPLGGDDPEYTAATRGAAAP---
MAV_4494|M.avium_104 DDAVLVMAGAYRSYAHHHPGRYSAFTRMPLGGDDPEYTAATRGAAAP---
MMAR_1010|M.marinum_M DDAVLVMAGAYRSYAHHHPGRYSAFTRMPPGGEDPEYAAATRAAAAP---
MUL_0762|M.ulcerans_Agy99 DDAVLVMAGAYRSYAHHHPGRYSAFTRMPPGGEDPEYAAATRAAAAP---
MAB_3852c|M.abscessus_ATCC_199 EDAILAMAGAYRSYAHHHPGRYAALTRMPFLDAVNRPTIDARELAKP---
MSMEG_0042|M.smegmatis_MC2_155 DHAVTRIAQAYLDFARANPAVYDAMFAR--ATNLRFAADDTPSPLTE---
Mflv_2110|M.gilvum_PYR-GCK VADFLMMGLAYHRFALANPQRYQLIFGAAAPASISRFRADITETGSATNR
MLBr_02568|M.leprae_Br4923 KDLLRNTIVSFLRYIDANRASWIVMYIQAMSLPAFAQTVREARDQIT---
. :: : : : :
TH_1893|M.thermoresistible__bu -------------VIAVLGSYGLEGEEAFYAALEFWSALHGFVLLEMTGV
Mvan_1278|M.vanbaalenii_PYR-1 -------------VISVLETFGLESESAFYAALEFWSAMHGFVLLEMTGV
Mb0700|M.bovis_AF2122/97 -------------VIAVLSSYGLDGEQAFYAALEFWSALHGFVLLEMTGV
Rv0681|M.tuberculosis_H37Rv -------------VIAVLSSYGLDGEQAFYAALEFWSALHGFVLLEMTGV
MAV_4494|M.avium_104 -------------VISVLSSYGLDGEEAFHAALEFWSALHGFVLLEMTGV
MMAR_1010|M.marinum_M -------------VIAVLSSYGLEGEQAFHAALEFWSALHGFVLLEMTGL
MUL_0762|M.ulcerans_Agy99 -------------VIAVLSSYGLEGEQAFHAALEFWSALHGFVLLEMTGL
MAB_3852c|M.abscessus_ATCC_199 -------------ATEAIGVYGLNEDTAFHAGVELWAAMHGFVMLEMTGF
MSMEG_0042|M.smegmatis_MC2_155 -------------AFAQLRDAVGRNEDADTLTEVLWAALHGLVMLGRTGR
Mflv_2110|M.gilvum_PYR-GCK AEWAASFGALHGAVHRMMAAGRIRRDDELAVAGRLWSINHGAVMLEMAGF
MLBr_02568|M.leprae_Br4923 ------------ELVAGLMRVGTRSSRPDHEYEIMAGALVGAGEAMATRL
: . : . * :
TH_1893|M.thermoresistible__bu MD--DVDSDKVFTDMVLRLAAGMEKRS---------------
Mvan_1278|M.vanbaalenii_PYR-1 MS--GIDTDAVFTDMVARLAAGMQLR----------------
Mb0700|M.bovis_AF2122/97 MD--DIDTDAVFTDMVLRLAAGMERRTTHGGTAST-------
Rv0681|M.tuberculosis_H37Rv MD--DIDTDAVFTDMVLRLAAGMERRTTHGGTAST-------
MAV_4494|M.avium_104 MD--DIDTDALFSDMVLRLAAGLERRTAHG------------
MMAR_1010|M.marinum_M ME--EIDTDAVFSDMVLRLAAGMEKRISHTGAESL-------
MUL_0762|M.ulcerans_Agy99 ME--EIDTDAVFSDMVLRLAAGMEKRISHTGAESL-------
MAB_3852c|M.abscessus_ATCC_199 MAGIEMDPDTAFAEMVHRFASGLAERK---------------
MSMEG_0042|M.smegmatis_MC2_155 LR------PDHETERLQLLVDRMSR-----------------
Mflv_2110|M.gilvum_PYR-GCK FGHEGHGLVHILGPLIIDTLVGMGDDRDAATLSMQTVVARLG
MLBr_02568|M.leprae_Br4923 TTG-DIAVDEAAELMINLFWHGLKGAPADRDFGPSIVAG---
: :