For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TLVLRYAARSDRGLVRANNEDSVYAGARLLALADGMGGHAAGEVASQLVIAALAHLDDDEPGGDLLSKLH QAVAEGNAAIAAHVEADPELDGMGTTLTAILFAGNRLGLVHIGDSRGYLLRDGELTQITKDDTFVQTLVD EGRITAEEAHSHPQRSLIMRALTGHEVEPTLIMREAKAGDRYLLCSDGLSDPVSQETIKEALQIEDVAES ADRLIELALRGGGPDNVTVVVADVVDYDYGQTQPILAGAVSGDDDQTAPPDTAAGRASAFNPRRNEPKRV IPQPAEPARKPRSRRRMIIAAVLVVLVVVAGLAIGREILRNNYYVAEHDGTVSIMRGVQGSFLGISLQEP YRLGCLNARNELSLIGANENRDNLGCRLMAVNDMRPSERAQVVAGLPGGTLDEAFAQIGELARSSLLPVC APPRPTTTTRAPAPPPAPPSSPAPGASGRGAGESPVPPSPSPTTTAAPAPSGAPGSPNPPPPASPSPTPS PTVTALPPPPPQPGTNCRAVS*
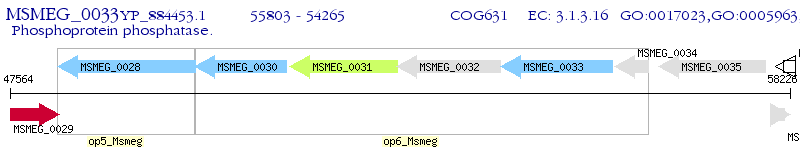
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0033 | - | - | 100% (512) | protein phosphatase 2C |
| M. smegmatis MC2 155 | MSMEG_1928 | - | 3e-29 | 37.76% (241) | regulatory protein |
| M. smegmatis MC2 155 | MSMEG_3618 | - | 1e-13 | 44.32% (88) | alanine and proline-rich secreted protein apa |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0018c | ppp | 0.0 | 71.21% (521) | serine/threonine phosphatase |
| M. gilvum PYR-GCK | Mflv_0808 | - | 0.0 | 78.79% (528) | protein phosphatase 2C domain-containing protein |
| M. tuberculosis H37Rv | Rv0018c | ppp | 0.0 | 71.40% (521) | serine/threonine phosphatase |
| M. leprae Br4923 | MLBr_00020 | - | 0.0 | 69.01% (513) | hypothetical protein MLBr_00020 |
| M. abscessus ATCC 19977 | MAB_0037c | - | 1e-180 | 65.14% (525) | serine/threonine phosphatase Ppp |
| M. marinum M | MMAR_0020 | pstP | 0.0 | 73.65% (520) | serine/threonine phosphatase PstP |
| M. avium 104 | MAV_0022 | - | 0.0 | 69.31% (518) | protein phosphatase 2C |
| M. thermoresistible (build 8) | TH_3554 | - | 2e-08 | 31.48% (108) | PUTATIVE - |
| M. ulcerans Agy99 | MUL_0022 | pstP | 0.0 | 73.46% (520) | serine/threonine phosphatase PstP |
| M. vanbaalenii PYR-1 | Mvan_0027 | - | 0.0 | 82.15% (521) | protein phosphatase 2C domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0808|M.gilvum_PYR-GCK ---MTLVLRYAARSDRGLVRANNEDSVYAGARLLALADGMGGHAAGEVAS
Mvan_0027|M.vanbaalenii_PYR-1 ---MTLVLRYAARSDRGLVRANNEDSVYAGARLLALADGMGGHAAGEVAS
MSMEG_0033|M.smegmatis_MC2_155 ---MTLVLRYAARSDRGLVRANNEDSVYAGARLLALADGMGGHAAGEVAS
MLBr_00020|M.leprae_Br4923 ---MTLVLRYAARSDRGLVRANNEDSVYAGARLLALADGMGGHAAGEVAS
MAV_0022|M.avium_104 ---MTLVLRYAARSDRGLVRSNNEDSVYAGARLLALADGMGGHAAGEVAS
Mb0018c|M.bovis_AF2122/97 MALVTLVLRYAARSDRGLVRANNEDSVYAGARLLALADGMGGHAAGEVAS
Rv0018c|M.tuberculosis_H37Rv MARVTLVLRYAARSDRGLVRANNEDSVYAGARLLALADGMGGHAAGEVAS
MMAR_0020|M.marinum_M ---MTLVLRYAARSDRGLVRANNEDSVYAGARLLALADGMGGHAAGEVAS
MUL_0022|M.ulcerans_Agy99 ---MTLVLRYAARSDRGLVRANNEDSVYAGARLLALADGMGGHAAGEVAS
MAB_0037c|M.abscessus_ATCC_199 ---MTLVLRYAARSDRGLVRSNNEDSVYAGARLLALADGMGGHAAGEVAS
TH_3554|M.thermoresistible__bu -------------------------------------------VAADAAA
.*.:.*:
Mflv_0808|M.gilvum_PYR-GCK QLVIAALAHLDDDEPGGDLLSKLDSAVREGNNAIASHVEADPELEGMGTT
Mvan_0027|M.vanbaalenii_PYR-1 QLVIAALAHLDDDEPGGDLLSKLDSAVREGNSAIAAHVEADPELDGMGTT
MSMEG_0033|M.smegmatis_MC2_155 QLVIAALAHLDDDEPGGDLLSKLHQAVAEGNAAIAAHVEADPELDGMGTT
MLBr_00020|M.leprae_Br4923 QLVIAALAHLDDDEPGGDLLGKLETAVRAGNLAIAAQVEMEPDFEGMGTT
MAV_0022|M.avium_104 QLVIAALAHLDDDEPGGDLLAKLDEAVRAGNAAIAAQVEAEPELEGMGTT
Mb0018c|M.bovis_AF2122/97 QLVIAALAHLDDDEPGGDLLAKLDAAVRAGNSAIAAQVEMEPDLEGMGTT
Rv0018c|M.tuberculosis_H37Rv QLVIAALAHLDDDEPGGDLLAKLDAAVRAGNSAIAAQVEMEPDLEGMGTT
MMAR_0020|M.marinum_M QLVIAALAHLDDDEPGGDLLAKLDAAVRSGNSAIAAQVEMEPELEGMGTT
MUL_0022|M.ulcerans_Agy99 QLVIAALAHLDDDEPGGDLLAKLDAAVRSGNSAIAAQVEMEPELEGMGTT
MAB_0037c|M.abscessus_ATCC_199 QLVIAALAPLDDDEPGGDLLDALADAVHAGNGAIADHVRAEPELDGMGTT
TH_3554|M.thermoresistible__bu PTAAAAAVPADPE--GSSTFRALAWSQEEG-------VDDVVPFTGDGAY
. ** . * : *.. : * : * * : * *:
Mflv_0808|M.gilvum_PYR-GCK LTAILFAGNRLGLVHIGDSRGYLLRDGELTQITKDDTFVQTLVDEGRITA
Mvan_0027|M.vanbaalenii_PYR-1 LTAILFAGNRLGLVHIGDSRGYLLRDGELTQITKDDTFVQTLVDEGRITA
MSMEG_0033|M.smegmatis_MC2_155 LTAILFAGNRLGLVHIGDSRGYLLRDGELTQITKDDTFVQTLVDEGRITA
MLBr_00020|M.leprae_Br4923 LTAILFAGNRLGLVHIGDSRGYLLRDGELTQITKDDTFVQTLVDEGRITA
MAV_0022|M.avium_104 LTAILFAGDRIGLVHIGDSRGYLLRDGELTQITKDDTFVQTLVDEGRITR
Mb0018c|M.bovis_AF2122/97 LTAILFAGNRLGLVHIGDSRGYLLRDGELTQITKDDTFVQTLVDEGRITP
Rv0018c|M.tuberculosis_H37Rv LTAILFAGNRLGLVHIGDSRGYLLRDGELTQITKDDTFVQTLVDEGRITP
MMAR_0020|M.marinum_M LTAILFAGNRLGLVHIGDSRGYLLRDGELTQITKDDTFVQTLVDEGRITA
MUL_0022|M.ulcerans_Agy99 LTAILFAGNRLGLVHIGDSRGYLLRDGELTQITKDDTFVQTLVDEGRITA
MAB_0037c|M.abscessus_ATCC_199 LTAFLFAGKRLGMVHIGDSRAYLMRDGELTQISKDDTFVQTLVDEGRITP
TH_3554|M.thermoresistible__bu PDGPFTEGGYTDGPFTGQAR------PEVQWVPEQ-------------PV
. : * . . *::* *: :.:: .
Mflv_0808|M.gilvum_PYR-GCK EEAHSHPQRSLIMRALTGHEVEPTLIMREARAGDRYLLCSDGLSDPVSQD
Mvan_0027|M.vanbaalenii_PYR-1 EEAHSHPQRSLIMRALTGHEVEPTLIMREARAGDRYLLCSDGLSDPVSHD
MSMEG_0033|M.smegmatis_MC2_155 EEAHSHPQRSLIMRALTGHEVEPTLIMREAKAGDRYLLCSDGLSDPVSQE
MLBr_00020|M.leprae_Br4923 EEAHSHPQRSLIMRALTGHEVDPTLTMREARAGDRYLLCSDGLSDPVSDE
MAV_0022|M.avium_104 EEAHSHPQRSLIMRALTGHEVEPTLTMREARAGDRYLLCSDGLSDPVSDE
Mb0018c|M.bovis_AF2122/97 EEAHSHPQRSLIMRALTGHEVEPTLTMREARAGDRYLLCSDGLSDPVSDE
Rv0018c|M.tuberculosis_H37Rv EEAHSHPQRSLIMRALTGHEVEPTLTMREARAGDRYLLCSDGLSDPVSDE
MMAR_0020|M.marinum_M EEAHSHPQRSLIMRALTGHEVEPTLTMREARAGDRYLLCSDGLSDPVSDE
MUL_0022|M.ulcerans_Agy99 EEAHSHPQRSLIMRALTGHEVEPTLTIREARAGDRYLLCSDGLSDPVSDE
MAB_0037c|M.abscessus_ATCC_199 EQAHNHPQRSLIMRALTGAPVEPTLIMREAHAGDRYLLCSDGLSDPVSHE
TH_3554|M.thermoresistible__bu EESGAWSRLPVALFSVAG------VAVLLALGGVAYGLTSVSEDEPD---
*:: .: .: : :::* : : * .* * * * . .:*
Mflv_0808|M.gilvum_PYR-GCK TIAEALQIDDVTESADRLIELALRGGGPDNVTVVVADVVDYDYGQTQPIL
Mvan_0027|M.vanbaalenii_PYR-1 TIAEALQIEDVAESADRLIELALRGGGPDNVTVVVADVVDYDYGQTQPIL
MSMEG_0033|M.smegmatis_MC2_155 TIKEALQIEDVAESADRLIELALRGGGPDNVTVVVADVVDYDYGQTQPIL
MLBr_00020|M.leprae_Br4923 TILEALQIPDVAESAYRLIELALRGGGPDNVTVVVADVIDYDYGQTQPIL
MAV_0022|M.avium_104 TILEALQIPDVAEAAYRLIELALRGGGPDNVTVVVADVVDYDYGQTQPIL
Mb0018c|M.bovis_AF2122/97 TILEALQIPEVAESAHRLIELALRGGGPDNVTVVVADVVDYDYGQTQPIL
Rv0018c|M.tuberculosis_H37Rv TILEALQIPEVAESAHRLIELALRGGGPDNVTVVVADVVDYDYGQTQPIL
MMAR_0020|M.marinum_M TIHEALQIPDVAESAYRLIELALRGGGPDNVTVVVADVVDYEYGQTQPIL
MUL_0022|M.ulcerans_Agy99 TIHEALQIPDVAESAYRLIELALRGGGPDNVTVVVADVVDYEYGQTQPIL
MAB_0037c|M.abscessus_ATCC_199 TIAEALEIPDVSAAADRLIELALRSGGPDNVTVVVADVVELDYGQTRPIV
TH_3554|M.thermoresistible__bu --------PPVTEXXX---QVLFAAGVVAVLVALIAAIVTLRDGDTP---
*: :: : .* :..::* :: *:*
Mflv_0808|M.gilvum_PYR-GCK AGAVSGDD-DQSAPPNTAAGRASAFNPKRNAAKRVVPQPE------EPPP
Mvan_0027|M.vanbaalenii_PYR-1 AGAVSGDD-DQSDPPNTAAGRASAFNPKRNAAKRVVPQPE------EPPP
MSMEG_0033|M.smegmatis_MC2_155 AGAVSGDD-DQTAPPDTAAGRASAFNPRRNEPKRVIPQPA------EPAR
MLBr_00020|M.leprae_Br4923 AGAVSGEESQQMALPNTAASRASAIVPRKEIAKSMAPQVD------KVHQ
MAV_0022|M.avium_104 AGAVSGDE-DQLTLPNTSAGRASAIRPRDESAKRVAPQPE------TPSR
Mb0018c|M.bovis_AF2122/97 AGAVSGDD-DQLTLPNTAAGRASAISQRKEIVKRVPPQAD------TFSR
Rv0018c|M.tuberculosis_H37Rv AGAVSGDD-DQLTLPNTAAGRASAISQRKEIVKRVPPQAD------TFSR
MMAR_0020|M.marinum_M AGAVSGDD-DQLTLPNTAAGRASAITARKEAVKRVPPQVE------TVHR
MUL_0022|M.ulcerans_Agy99 AGAVSGDD-DQLTLPNTAAGRASAITARKEAVKRVPPQVE------TVHR
MAB_0037c|M.abscessus_ATCC_199 AGAVSTDADTDTPPPNTAAGRAAAIQRALKEPEPAEDESDSETTDSENSK
TH_3554|M.thermoresistible__bu APASTTEP----TEPTTTEPAGTVVTEAPPPAEPAAPQPP----------
* * : : * *: .:.. : :
Mflv_0808|M.gilvum_PYR-GCK RPRSRRRPIIAATVLVLVVLAGLAVGREIVRNNYYVSENDGTVSIMRGVQ
Mvan_0027|M.vanbaalenii_PYR-1 RPHSRRRMIIAAAVLVLVVLTGLAVGREIVRNNYYVSEHNGTVSIMRGVQ
MSMEG_0033|M.smegmatis_MC2_155 KPRSRRRMIIAAVLVVLVVVAGLAIGREILRNNYYVAEHDGTVSIMRGVQ
MLBr_00020|M.leprae_Br4923 QRWSQRQTGITLTLLVLLVLAGVAIGRWVIRDNYYIAEYSGKVSIVRGIQ
MAV_0022|M.avium_104 PRWSRRRLFIVIALAVMLVLAGLTVGWWVIQRNYYVAEYNGRISIVRGIQ
Mb0018c|M.bovis_AF2122/97 PRWSGRRLAFVVALVTVLMTAGLLIGRAIIRSNYYVADYAGSVSIMRGIQ
Rv0018c|M.tuberculosis_H37Rv PRWSGRRLAFVVALVTVLMTAGLLIGRAIIRSNYYVADYAGSVSIMRGIQ
MMAR_0020|M.marinum_M PRWSWRRMALVATLVVLLVVAGLFISRAVIRNNYYVAEFNGTVSIMRGIQ
MUL_0022|M.ulcerans_Agy99 PRWSWRRMALVAELVVLLVVAGLFISRAVIRNNYYVAEFDGTVSIMRGIQ
MAB_0037c|M.abscessus_ATCC_199 PKKRRLRWLIAAVLVVLIALAGLAIARRVVLNNYYVSAHEGNVSIMQGVQ
TH_3554|M.thermoresistible__bu ----------------------------------------------RTVY
: :
Mflv_0808|M.gilvum_PYR-GCK GSFLGVSLQEPYLLGCLNNRNELSLISADQSGEPRDCRLLGVDDMRPSER
Mvan_0027|M.vanbaalenii_PYR-1 GSFLGVSLQEPYLLGCLNARNELSLISAGQSQDSLDCRLLGVGDMRPSER
MSMEG_0033|M.smegmatis_MC2_155 GSFLGISLQEPYRLGCLNARNELSLIGANENRDNLGCRLMAVNDMRPSER
MLBr_00020|M.leprae_Br4923 GSLLGMPLHEPYLVGCLNTRNELSLISYGQSGGHLNCQVMTLRDLRPSGG
MAV_0022|M.avium_104 GSLLGVPLQQPYLVGCLNARNELSLISYGQSGGS-NCQLMTLQDLRRPGQ
Mb0018c|M.bovis_AF2122/97 GSLLGMSLHQPYLMGCLSPRNELSQISYGQSGGPLDCHLMKLEDLRPPER
Rv0018c|M.tuberculosis_H37Rv GSLLGMSLHQPYLMGCLSPRNELSQISYGQSGGPLDCHLMKLEDLRPPER
MMAR_0020|M.marinum_M GSFLGMSLHEPYLMGCLNARNELSQISYQQSGEHLDCNLMKLDDLRPPER
MUL_0022|M.ulcerans_Agy99 GSFLGMSLHEPYLMGCLNARNELSQISYQQSGEHLDCNLMKLDDLRPPER
MAB_0037c|M.abscessus_ATCC_199 GSLLGYRLQRPDLVGCLDSRGDMSLVSYGEAASNLSCKYLKVSDLRPSER
TH_3554|M.thermoresistible__bu ETVPQTQAPPP---------------------------------------
:. *
Mflv_0808|M.gilvum_PYR-GCK AQVIAGLPSGSLDQAIGQIEELSRSSVLPVCATHTTPTPSPRPAPSPG-A
Mvan_0027|M.vanbaalenii_PYR-1 AQVIAGLPSGSLDEAIGQIEELARSSVLPVCAPPAPPSTTRPPAPSPRPA
MSMEG_0033|M.smegmatis_MC2_155 AQVVAGLPGGTLDEAFAQIGELARSSLLPVCAPPRPTTTTRAPAPPPAPP
MLBr_00020|M.leprae_Br4923 AQVQAGLPGGNLDQAESQLRKLLADSLLPTCPPPRATSPPGPFTMPTDSG
MAV_0022|M.avium_104 VQVQTGLPGGSLDQAESQLRQLLAEYLLPLCPPPRATSPPG--AQATRSP
Mb0018c|M.bovis_AF2122/97 AQVRAGLPAGTLDDAIGQLRELAANSLLPPCPAPRATSPPGRPAPPTTSE
Rv0018c|M.tuberculosis_H37Rv AQVRAGLPAGTLDDAIGQLRELAANSLLPPCPAPRATSPPGRPAPPTTSE
MMAR_0020|M.marinum_M AQVQAGLPAGSLDNAIGQLRELAANSLLPPCPAPRATSPPESSAPSTASE
MUL_0022|M.ulcerans_Agy99 AQVQAGLPAGSLDNAIGQLRELAANSLLPPCPAPRATSPPEPSAPSTASE
MAB_0037c|M.abscessus_ATCC_199 QQVETGLPGGTLDEAIGQLRQLSKN-ALPPCPPPAPKTPPP-PAPTAAGR
TH_3554|M.thermoresistible__bu ----------------------------PAEAPPPAQEPPPAEEPPPAEE
* .. . .. ..
Mflv_0808|M.gilvum_PYR-GCK SLPRPAPPASTPTTTPPESPAASPDPRTGTPAPVPPRTSSAPSPSGS--A
Mvan_0027|M.vanbaalenii_PYR-1 PAPTSTAPQTTGPSTTPRASGPAPAPETPRTVTTTAPSPASPAPVPP--A
MSMEG_0033|M.smegmatis_MC2_155 SSPAPGASG-RGAGESPVPPSPSP---TTTAAPAPSGAPGSPNPPPP--A
MLBr_00020|M.leprae_Br4923 TP-QPTNTASPTST---TPPTITSSSGTAPTNISPTSSPASTAPTTP---
MAV_0022|M.avium_104 VP-ETGGPASP-------APPTTSASPTPSTNATPG--PASSSPAGP---
Mb0018c|M.bovis_AF2122/97 TT-EPNVTSSPAA-----PSPTTSASAPTGTTPAIPTSASPAAPASP--P
Rv0018c|M.tuberculosis_H37Rv TT-EPNVTSSPAS-----PSPTTSAPAPTGTTPAIPTSASPAAPASP--P
MMAR_0020|M.marinum_M TPGQPSVTSSPASTTPPTPSASTTPTTTSGSTAATSTPPTGTSPAAPTSP
MUL_0022|M.ulcerans_Agy99 TPGQPSVTSSPASTTTPTPSASTTPTTTSGSTAATSTPPAGTSPAAPTSP
MAB_0037c|M.abscessus_ATCC_199 TP-----------------TTTSGTPTTTPSKPSPTTSAGPGASTTP---
TH_3554|M.thermoresistible__bu PP--------------------PATEEPPPTTTRPRWLPTLPRPTLP---
. . . . . .
Mflv_0808|M.gilvum_PYR-GCK PPPPPSPSPTVTALPPPPPEPGTNCREVS
Mvan_0027|M.vanbaalenii_PYR-1 SPTPPPPPPTVTALPPPPPEPGTNCREVS
MSMEG_0033|M.smegmatis_MC2_155 SPSP-TPSPTVTALPPPPPQPGTNCRAVS
MLBr_00020|M.leprae_Br4923 ----VGTSQTVTILPPPPPQPGIDCRAVA
MAV_0022|M.avium_104 ----TTTSQTLTALPGPPLQPGIDCRTVA
Mb0018c|M.bovis_AF2122/97 TPWPVTSSPTMAALPPPPPQPGIDCRAAA
Rv0018c|M.tuberculosis_H37Rv TPWPVTSSPTMAALPPPPPQPGIDCRAAA
MMAR_0020|M.marinum_M TPTLTTSSPNVTALPPPPPQPGIDCRAVA
MUL_0022|M.ulcerans_Agy99 TPTSTTSSPNVTALPPPPPQPGIDCRAVA
MAB_0037c|M.abscessus_ATCC_199 ---TTTSTPAPAPLPAPPQEPGVTCRTVS
TH_3554|M.thermoresistible__bu ------TLPTIPGLPAPPAPPAEAP----
. . ** ** *.