For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MASVLSAATATDQGPVRENNQDACLADGILYAVADGFGARGHHASATALKTLSAGFAAAPDRDGLLEAVQ QANLRVFELLGDEPTVSGTTLTAVAVFEPGQGGPLVVNIGDSPLYRIRDGHMEQLTDDHSVAGELVRMGE ITRHEARWHPQRHLLTRALGIGPHIGPDVFGIDCGPGDRLLISSDGLFAAADEALIVDAATSPDPQVAVR RLVEVANDAGGSDNTTVVVIDLG
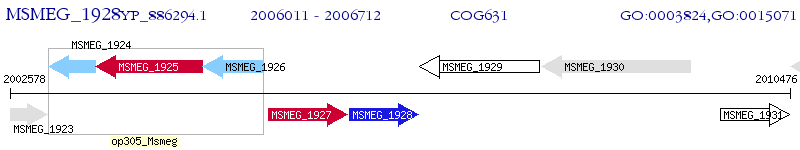
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1928 | - | - | 100% (233) | regulatory protein |
| M. smegmatis MC2 155 | MSMEG_0033 | - | 1e-29 | 37.76% (241) | protein phosphatase 2C |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0018c | ppp | 7e-30 | 38.24% (238) | serine/threonine phosphatase |
| M. gilvum PYR-GCK | Mflv_0808 | - | 3e-29 | 37.82% (238) | protein phosphatase 2C domain-containing protein |
| M. tuberculosis H37Rv | Rv0018c | ppp | 7e-30 | 38.24% (238) | serine/threonine phosphatase |
| M. leprae Br4923 | MLBr_00020 | - | 1e-31 | 39.92% (238) | hypothetical protein MLBr_00020 |
| M. abscessus ATCC 19977 | MAB_0037c | - | 6e-31 | 38.24% (238) | serine/threonine phosphatase Ppp |
| M. marinum M | MMAR_0020 | pstP | 1e-29 | 38.66% (238) | serine/threonine phosphatase PstP |
| M. avium 104 | MAV_0022 | - | 4e-32 | 39.92% (238) | protein phosphatase 2C |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_0022 | pstP | 9e-30 | 38.66% (238) | serine/threonine phosphatase PstP |
| M. vanbaalenii PYR-1 | Mvan_0027 | - | 5e-29 | 37.82% (238) | protein phosphatase 2C domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_00020|M.leprae_Br4923 ---MTLVLRYAARSDRGLVRANNEDSVYAGARLLALADGMGGHAAGEVAS
MAV_0022|M.avium_104 ---MTLVLRYAARSDRGLVRSNNEDSVYAGARLLALADGMGGHAAGEVAS
Mb0018c|M.bovis_AF2122/97 MALVTLVLRYAARSDRGLVRANNEDSVYAGARLLALADGMGGHAAGEVAS
Rv0018c|M.tuberculosis_H37Rv MARVTLVLRYAARSDRGLVRANNEDSVYAGARLLALADGMGGHAAGEVAS
MMAR_0020|M.marinum_M ---MTLVLRYAARSDRGLVRANNEDSVYAGARLLALADGMGGHAAGEVAS
MUL_0022|M.ulcerans_Agy99 ---MTLVLRYAARSDRGLVRANNEDSVYAGARLLALADGMGGHAAGEVAS
Mflv_0808|M.gilvum_PYR-GCK ---MTLVLRYAARSDRGLVRANNEDSVYAGARLLALADGMGGHAAGEVAS
Mvan_0027|M.vanbaalenii_PYR-1 ---MTLVLRYAARSDRGLVRANNEDSVYAGARLLALADGMGGHAAGEVAS
MAB_0037c|M.abscessus_ATCC_199 ---MTLVLRYAARSDRGLVRSNNEDSVYAGARLLALADGMGGHAAGEVAS
MSMEG_1928|M.smegmatis_MC2_155 ---MASVLSAATATDQGPVRENNQDACLADGILYAVADGFG--ARGHHAS
:: ** *: :*:* ** **:*: *.. * *:***:* * *. **
MLBr_00020|M.leprae_Br4923 QLVIAALAHLDDDEPGGDLLGKLETAVRAGNLAIAAQVEMEPDFEGMGTT
MAV_0022|M.avium_104 QLVIAALAHLDDDEPGGDLLAKLDEAVRAGNAAIAAQVEAEPELEGMGTT
Mb0018c|M.bovis_AF2122/97 QLVIAALAHLDDDEPGGDLLAKLDAAVRAGNSAIAAQVEMEPDLEGMGTT
Rv0018c|M.tuberculosis_H37Rv QLVIAALAHLDDDEPGGDLLAKLDAAVRAGNSAIAAQVEMEPDLEGMGTT
MMAR_0020|M.marinum_M QLVIAALAHLDDDEPGGDLLAKLDAAVRSGNSAIAAQVEMEPELEGMGTT
MUL_0022|M.ulcerans_Agy99 QLVIAALAHLDDDEPGGDLLAKLDAAVRSGNSAIAAQVEMEPELEGMGTT
Mflv_0808|M.gilvum_PYR-GCK QLVIAALAHLDDDEPGGDLLSKLDSAVREGNNAIASHVEADPELEGMGTT
Mvan_0027|M.vanbaalenii_PYR-1 QLVIAALAHLDDDEPGGDLLSKLDSAVREGNSAIAAHVEADPELDGMGTT
MAB_0037c|M.abscessus_ATCC_199 QLVIAALAPLDDDEPGGDLLDALADAVHAGNGAIADHVRAEPELDGMGTT
MSMEG_1928|M.smegmatis_MC2_155 ATALKTLSAGFAAAPDRD---GLLEAVQQANLRVFELLGDEPTVS--GTT
.: :*: *. * * **: .* : : :* .. ***
MLBr_00020|M.leprae_Br4923 LTAILFAGNRLG---LVHIGDSRGYLLRDGELTQITKDDTFVQTLVDEGR
MAV_0022|M.avium_104 LTAILFAGDRIG---LVHIGDSRGYLLRDGELTQITKDDTFVQTLVDEGR
Mb0018c|M.bovis_AF2122/97 LTAILFAGNRLG---LVHIGDSRGYLLRDGELTQITKDDTFVQTLVDEGR
Rv0018c|M.tuberculosis_H37Rv LTAILFAGNRLG---LVHIGDSRGYLLRDGELTQITKDDTFVQTLVDEGR
MMAR_0020|M.marinum_M LTAILFAGNRLG---LVHIGDSRGYLLRDGELTQITKDDTFVQTLVDEGR
MUL_0022|M.ulcerans_Agy99 LTAILFAGNRLG---LVHIGDSRGYLLRDGELTQITKDDTFVQTLVDEGR
Mflv_0808|M.gilvum_PYR-GCK LTAILFAGNRLG---LVHIGDSRGYLLRDGELTQITKDDTFVQTLVDEGR
Mvan_0027|M.vanbaalenii_PYR-1 LTAILFAGNRLG---LVHIGDSRGYLLRDGELTQITKDDTFVQTLVDEGR
MAB_0037c|M.abscessus_ATCC_199 LTAFLFAGKRLG---MVHIGDSRAYLMRDGELTQISKDDTFVQTLVDEGR
MSMEG_1928|M.smegmatis_MC2_155 LTAVAVFEPGQGGPLVVNIGDSPLYRIRDGHMEQLTDDHSVAGELVRMGE
***. . * :*:**** * :***.: *::.*.:.. ** *.
MLBr_00020|M.leprae_Br4923 ITAEEAHSHPQRSLIMRAL-TGHEVDPTLTMREARAGDRYLLCSDGLSDP
MAV_0022|M.avium_104 ITREEAHSHPQRSLIMRAL-TGHEVEPTLTMREARAGDRYLLCSDGLSDP
Mb0018c|M.bovis_AF2122/97 ITPEEAHSHPQRSLIMRAL-TGHEVEPTLTMREARAGDRYLLCSDGLSDP
Rv0018c|M.tuberculosis_H37Rv ITPEEAHSHPQRSLIMRAL-TGHEVEPTLTMREARAGDRYLLCSDGLSDP
MMAR_0020|M.marinum_M ITAEEAHSHPQRSLIMRAL-TGHEVEPTLTMREARAGDRYLLCSDGLSDP
MUL_0022|M.ulcerans_Agy99 ITAEEAHSHPQRSLIMRAL-TGHEVEPTLTIREARAGDRYLLCSDGLSDP
Mflv_0808|M.gilvum_PYR-GCK ITAEEAHSHPQRSLIMRAL-TGHEVEPTLIMREARAGDRYLLCSDGLSDP
Mvan_0027|M.vanbaalenii_PYR-1 ITAEEAHSHPQRSLIMRAL-TGHEVEPTLIMREARAGDRYLLCSDGLSDP
MAB_0037c|M.abscessus_ATCC_199 ITPEQAHNHPQRSLIMRAL-TGAPVEPTLIMREAHAGDRYLLCSDGLSDP
MSMEG_1928|M.smegmatis_MC2_155 ITRHEARWHPQRHLLTRALGIGPHIGPDVFGIDCGPGDRLLISSDGLFAA
** .:*: **** *: *** * : * : :. .*** *:.**** .
MLBr_00020|M.leprae_Br4923 VSDETILEALQIPDVAESAYRLIELALRGGGPDNVTVVVADVIDYDYGQT
MAV_0022|M.avium_104 VSDETILEALQIPDVAEAAYRLIELALRGGGPDNVTVVVADVVDYDYGQT
Mb0018c|M.bovis_AF2122/97 VSDETILEALQIPEVAESAHRLIELALRGGGPDNVTVVVADVVDYDYGQT
Rv0018c|M.tuberculosis_H37Rv VSDETILEALQIPEVAESAHRLIELALRGGGPDNVTVVVADVVDYDYGQT
MMAR_0020|M.marinum_M VSDETIHEALQIPDVAESAYRLIELALRGGGPDNVTVVVADVVDYEYGQT
MUL_0022|M.ulcerans_Agy99 VSDETIHEALQIPDVAESAYRLIELALRGGGPDNVTVVVADVVDYEYGQT
Mflv_0808|M.gilvum_PYR-GCK VSQDTIAEALQIDDVTESADRLIELALRGGGPDNVTVVVADVVDYDYGQT
Mvan_0027|M.vanbaalenii_PYR-1 VSHDTIAEALQIEDVAESADRLIELALRGGGPDNVTVVVADVVDYDYGQT
MAB_0037c|M.abscessus_ATCC_199 VSHETIAEALEIPDVSAAADRLIELALRSGGPDNVTVVVADVVELDYGQT
MSMEG_1928|M.smegmatis_MC2_155 ADEALIVDAATSPDPQVAVRRLVEVANDAGGSDNTTVVVIDLG-------
... * :* : :. **:*:* .**.**.**** *:
MLBr_00020|M.leprae_Br4923 QPILAGAVSGEESQQMALPNTAASRASAIVPRKEIAKSMAPQVD------
MAV_0022|M.avium_104 QPILAGAVSGDE-DQLTLPNTSAGRASAIRPRDESAKRVAPQPE------
Mb0018c|M.bovis_AF2122/97 QPILAGAVSGDD-DQLTLPNTAAGRASAISQRKEIVKRVPPQAD------
Rv0018c|M.tuberculosis_H37Rv QPILAGAVSGDD-DQLTLPNTAAGRASAISQRKEIVKRVPPQAD------
MMAR_0020|M.marinum_M QPILAGAVSGDD-DQLTLPNTAAGRASAITARKEAVKRVPPQVE------
MUL_0022|M.ulcerans_Agy99 QPILAGAVSGDD-DQLTLPNTAAGRASAITARKEAVKRVPPQVE------
Mflv_0808|M.gilvum_PYR-GCK QPILAGAVSGDD-DQSAPPNTAAGRASAFNPKRNAAKRVVPQPE------
Mvan_0027|M.vanbaalenii_PYR-1 QPILAGAVSGDD-DQSDPPNTAAGRASAFNPKRNAAKRVVPQPE------
MAB_0037c|M.abscessus_ATCC_199 RPIVAGAVSTDADTDTPPPNTAAGRAAAIQRALKEPEPAEDESDSETTDS
MSMEG_1928|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00020|M.leprae_Br4923 KVHQQRWSQRQTGITLTLLVLLVLAGVAIGRWVIRDNYYIAEYSGKVSIV
MAV_0022|M.avium_104 TPSRPRWSRRRLFIVIALAVMLVLAGLTVGWWVIQRNYYVAEYNGRISIV
Mb0018c|M.bovis_AF2122/97 TFSRPRWSGRRLAFVVALVTVLMTAGLLIGRAIIRSNYYVADYAGSVSIM
Rv0018c|M.tuberculosis_H37Rv TFSRPRWSGRRLAFVVALVTVLMTAGLLIGRAIIRSNYYVADYAGSVSIM
MMAR_0020|M.marinum_M TVHRPRWSWRRMALVATLVVLLVVAGLFISRAVIRNNYYVAEFNGTVSIM
MUL_0022|M.ulcerans_Agy99 TVHRPRWSWRRMALVAELVVLLVVAGLFISRAVIRNNYYVAEFDGTVSIM
Mflv_0808|M.gilvum_PYR-GCK EPPPRPRSRRRPIIAATVLVLVVLAGLAVGREIVRNNYYVSENDGTVSIM
Mvan_0027|M.vanbaalenii_PYR-1 EPPPRPHSRRRMIIAAAVLVLVVLTGLAVGREIVRNNYYVSEHNGTVSIM
MAB_0037c|M.abscessus_ATCC_199 ENSKPKKRRLRWLIAAVLVVLIALAGLAIARRVVLNNYYVSAHEGNVSIM
MSMEG_1928|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00020|M.leprae_Br4923 RGIQGSLLGMPLHEPYLVGCLNTRNELSLISYGQSGGHLNCQVMTLRDLR
MAV_0022|M.avium_104 RGIQGSLLGVPLQQPYLVGCLNARNELSLISYGQSGGS-NCQLMTLQDLR
Mb0018c|M.bovis_AF2122/97 RGIQGSLLGMSLHQPYLMGCLSPRNELSQISYGQSGGPLDCHLMKLEDLR
Rv0018c|M.tuberculosis_H37Rv RGIQGSLLGMSLHQPYLMGCLSPRNELSQISYGQSGGPLDCHLMKLEDLR
MMAR_0020|M.marinum_M RGIQGSFLGMSLHEPYLMGCLNARNELSQISYQQSGEHLDCNLMKLDDLR
MUL_0022|M.ulcerans_Agy99 RGIQGSFLGMSLHEPYLMGCLNARNELSQISYQQSGEHLDCNLMKLDDLR
Mflv_0808|M.gilvum_PYR-GCK RGVQGSFLGVSLQEPYLLGCLNNRNELSLISADQSGEPRDCRLLGVDDMR
Mvan_0027|M.vanbaalenii_PYR-1 RGVQGSFLGVSLQEPYLLGCLNARNELSLISAGQSQDSLDCRLLGVGDMR
MAB_0037c|M.abscessus_ATCC_199 QGVQGSLLGYRLQRPDLVGCLDSRGDMSLVSYGEAASNLSCKYLKVSDLR
MSMEG_1928|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00020|M.leprae_Br4923 PSGGAQVQAGLPGGNLDQAESQLRKLLADSLLPTCPPPRATSPPGPFTMP
MAV_0022|M.avium_104 RPGQVQVQTGLPGGSLDQAESQLRQLLAEYLLPLCPPPRATSPPG--AQA
Mb0018c|M.bovis_AF2122/97 PPERAQVRAGLPAGTLDDAIGQLRELAANSLLPPCPAPRATSPPGRPAPP
Rv0018c|M.tuberculosis_H37Rv PPERAQVRAGLPAGTLDDAIGQLRELAANSLLPPCPAPRATSPPGRPAPP
MMAR_0020|M.marinum_M PPERAQVQAGLPAGSLDNAIGQLRELAANSLLPPCPAPRATSPPESSAPS
MUL_0022|M.ulcerans_Agy99 PPERAQVQAGLPAGSLDNAIGQLRELAANSLLPPCPAPRATSPPEPSAPS
Mflv_0808|M.gilvum_PYR-GCK PSERAQVIAGLPSGSLDQAIGQIEELSRSSVLPVCATHTTPTPSPRPAPS
Mvan_0027|M.vanbaalenii_PYR-1 PSERAQVIAGLPSGSLDEAIGQIEELARSSVLPVCAPPAPPSTTRPPAPS
MAB_0037c|M.abscessus_ATCC_199 PSERQQVETGLPGGTLDEAIGQLRQLSKN-ALPPCPPP-APKTPPPPAPT
MSMEG_1928|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00020|M.leprae_Br4923 TDSGTP-QPTNTASPTST---TPPTITSSSGTAPTNISPTSSPASTAPTT
MAV_0022|M.avium_104 TRSPVP-ETGGPASP-------APPTTSASPTPSTNATPG--PASSSPAG
Mb0018c|M.bovis_AF2122/97 TTSETT-EPNVTSSPAA-----PSPTTSASAPTGTTPAIPTSASPAAPAS
Rv0018c|M.tuberculosis_H37Rv TTSETT-EPNVTSSPAS-----PSPTTSAPAPTGTTPAIPTSASPAAPAS
MMAR_0020|M.marinum_M TASETPGQPSVTSSPASTTPPTPSASTTPTTTSGSTAATSTPPTGTSPAA
MUL_0022|M.ulcerans_Agy99 TASETPGQPSVTSSPASTTTPTPSASTTPTTTSGSTAATSTPPAGTSPAA
Mflv_0808|M.gilvum_PYR-GCK PG-ASLPRPAPPASTPTTTPPESPAASPDPRTGTPAPVPPRTSSAPSPSG
Mvan_0027|M.vanbaalenii_PYR-1 PRPAPAPTSTAPQTTGPSTTPRASGPAPAPETPRTVTTTAPSPASPAPVP
MAB_0037c|M.abscessus_ATCC_199 AAGRTP-----------------TTTSGTPTTTPSKPSPTTSAGPGASTT
MSMEG_1928|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00020|M.leprae_Br4923 P-------VGTSQTVTILPPPPPQPGIDCRAVA
MAV_0022|M.avium_104 P-------TTTSQTLTALPGPPLQPGIDCRTVA
Mb0018c|M.bovis_AF2122/97 P--PTPWPVTSSPTMAALPPPPPQPGIDCRAAA
Rv0018c|M.tuberculosis_H37Rv P--PTPWPVTSSPTMAALPPPPPQPGIDCRAAA
MMAR_0020|M.marinum_M PTSPTPTLTTSSPNVTALPPPPPQPGIDCRAVA
MUL_0022|M.ulcerans_Agy99 PTSPTPTSTTSSPNVTALPPPPPQPGIDCRAVA
Mflv_0808|M.gilvum_PYR-GCK S--APPPPPSPSPTVTALPPPPPEPGTNCREVS
Mvan_0027|M.vanbaalenii_PYR-1 P--ASPTPPPPPPTVTALPPPPPEPGTNCREVS
MAB_0037c|M.abscessus_ATCC_199 P------TTTSTPAPAPLPAPPQEPGVTCRTVS
MSMEG_1928|M.smegmatis_MC2_155 ---------------------------------