For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTEPRMDWDSAYRQDVRPPWSIGAPQPELAALIEGGKVRSEVLDAGCGEAELSLALAAQGYSVVGLDASA TAIATASAAAAERGLTTASFAAADVTAFDGYDGRFNTVMDSGLLHVAGPRELVRGERESFIGGYRRQHSG QAVGSPAMNAADDGCTRASSRR
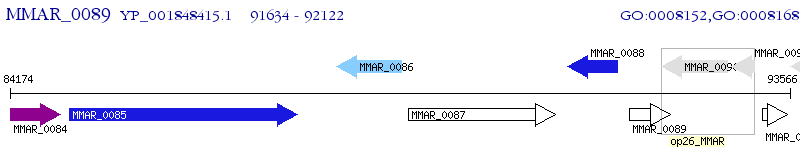
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_0089 | - | - | 100% (162) | hypothetical protein MMAR_0089 |
| M. marinum M | MMAR_5210 | - | 3e-36 | 61.79% (123) | hypothetical protein MMAR_5210 |
| M. marinum M | MMAR_0909 | - | 4e-34 | 55.47% (137) | methyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3725 | - | 1e-37 | 63.41% (123) | hypothetical protein Mb3725 |
| M. gilvum PYR-GCK | Mflv_1324 | - | 4e-38 | 68.70% (115) | methyltransferase type 11 |
| M. tuberculosis H37Rv | Rv3699 | - | 1e-37 | 63.41% (123) | hypothetical protein Rv3699 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0961c | - | 8e-35 | 61.40% (114) | hypothetical protein MAB_0961c |
| M. avium 104 | MAV_0404 | - | 2e-41 | 71.05% (114) | thiopurine S-methyltransferase (tpmt) superfamily protein |
| M. smegmatis MC2 155 | MSMEG_6235 | - | 1e-37 | 63.03% (119) | thiopurine S-methyltransferase (tpmt) superfamily protein |
| M. thermoresistible (build 8) | TH_3824 | - | 3e-41 | 67.80% (118) | PUTATIVE thiopurine S-methyltransferase (tpmt) superfamily |
| M. ulcerans Agy99 | MUL_0662 | - | 1e-32 | 59.17% (120) | methyltransferase |
| M. vanbaalenii PYR-1 | Mvan_5471 | - | 1e-36 | 66.09% (115) | methyltransferase type 11 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3725|M.bovis_AF2122/97 ---MTDEVMDWDSAYREQGA-FEG--PPPWNIGEPQPELATLIAA-----
Rv3699|M.tuberculosis_H37Rv ---MTDEVMDWDSAYREQGA-FEG--PPPWNIGEPQPELATLIAA-----
MAV_0404|M.avium_104 --MSSDQVMDWDSAYREQAH-FEG--PPPWNIGEPQPELAALIEQ-----
MSMEG_6235|M.smegmatis_MC2_155 MGVMTSHVLDWDGAYRGDTG-FQG--PPPWNIGEPQPELAALHRA-----
Mflv_1324|M.gilvum_PYR-GCK ----MAEVMDWDSAYRGEGE-FEG--EPPWNIGEPQPELAALWRD-----
Mvan_5471|M.vanbaalenii_PYR-1 -MAHMTDVMDWDSAYRGEGD-FEG--EPPWNIGEPQPELAALWRD-----
MAB_0961c|M.abscessus_ATCC_199 --MTSNELPDWDATYQGKGELFQG--EPPWNIGEPQPELAALIDA-----
MUL_0662|M.ulcerans_Agy99 --MTESLDLQFATAYRGESAEFGAGIRPPWSLGEPQPELATLIEQ-----
MMAR_0089|M.marinum_M ---MTEPRMDWDSAYRQDVR-------PPWSIGAPQPELAALIEG-----
TH_3824|M.thermoresistible__bu --------MEWDDMYRRGVP-------PPWSIGAPQPELAALLDAGPAQC
:: *: ***.:* ******:*
Mb3725|M.bovis_AF2122/97 ---GKVRSDVLDAGCGYAELSLALAADGYTVVGIDLTPTAVAAATKAAEE
Rv3699|M.tuberculosis_H37Rv ---GKVRSDVLDAGCGYAELSLALAADGYTVVGIDLTPTAVAAATKAAEE
MAV_0404|M.avium_104 ---GKFRSDVLDAGCGCAELSLALAARGYTVVGIDLTPTAVAAATRAAAE
MSMEG_6235|M.smegmatis_MC2_155 ---GRFRSDVLDAGCGHAELSLALAADGYTVVGMDLSPSAIAAANRAAQE
Mflv_1324|M.gilvum_PYR-GCK ---GRFRSEVLDAGCGHAELSLALAADGYTVTGVDISPTAMAAASAAAAE
Mvan_5471|M.vanbaalenii_PYR-1 ---GKFRSEVLDAGCGHAELSLALAADGYTVTGVDLSPTAVAAATAASAE
MAB_0961c|M.abscessus_ATCC_199 ---GKFHGTVLDVGCGHAETSLRLAALGHTTVGLDLSPTAIEAARAAAAE
MUL_0662|M.ulcerans_Agy99 ---GKVHGEVLDAGCGEAALAMALAGRGHPVVGLDISPTAVELAGREAAR
MMAR_0089|M.marinum_M ---GKVRSEVLDAGCGEAELSLALAAQGYSVVGLDASATAIATASAAAAE
TH_3824|M.thermoresistible__bu PVQGTVRGEVLDAGCGEAALSLELAARGHTVVGLDCSAAAVAAATATAAE
* .:. ***.*** * :: **. *:...*:* :.:*: * : .
Mb3725|M.bovis_AF2122/97 RGLTT-ASFVQADITEFAAYPAGSAGRFSTVIDSTLFHSLPVDSRDRYLS
Rv3699|M.tuberculosis_H37Rv RGLTT-ASFVQADITEFAAYPAGSAGRFSTVIDSTLFHSLPVDSRDRYLS
MAV_0404|M.avium_104 RGLTT-ASFVQADITSFTGY----DGRFATVVDSTLFHSLPVEGRDGYLR
MSMEG_6235|M.smegmatis_MC2_155 RNLAT-ASFVQGDITSFTGY----DGRFSTVIDSTLFHSLPVEGREGYLQ
Mflv_1324|M.gilvum_PYR-GCK RGLSEKASFVAADITALSGF----DGRFSTVVDSTLFHSLPVESRAGYLE
Mvan_5471|M.vanbaalenii_PYR-1 RGLSERTSFVAADITTLTGF----DGRFATVVDSTLFHSLPVESRDGYLQ
MAB_0961c|M.abscessus_ATCC_199 RGLTN-ATFEVADITDFSGY----DGRFNTIVDSTLFHSIPVEAREAYLQ
MUL_0662|M.ulcerans_Agy99 PGLTN-ASFAVADITDFASYPPESAGRFNTIMDSTLFHSMPVELREGCQR
MMAR_0089|M.marinum_M RGLTT-ASFAAADVTAFDGY----DGRFNTVMDSGLLHVA----------
TH_3824|M.thermoresistible__bu RGLTT-ATFAQADLTEFGGY----DGRFSTIIDSGLLHALPVDRRQAYLR
.*: ::* .*:* : .: *** *::** *:*
Mb3725|M.bovis_AF2122/97 SVHRAAAPGASYYVLVFAKGAFPAELEVKPNEVDEDELRAAVSKYWKIDE
Rv3699|M.tuberculosis_H37Rv SVHRAAAPGASYYVLVFAKGAFPAELEVKPNEVDEDELRAAVSKYWKIDE
MAV_0404|M.avium_104 SIHRAAAAGASLFILVFAKGAFPAEMQPKPNEVDEDELRAAVGKYWVIDE
MSMEG_6235|M.smegmatis_MC2_155 SIHRASAPGARYFVLVFAKGAFPADLEVKPNEVTEEELRTTVGKYWVVDD
Mflv_1324|M.gilvum_PYR-GCK SVHRAARPGAGFFVLVFAKGAFPADTEQGPNEVSEIELREAVSPYWTIDD
Mvan_5471|M.vanbaalenii_PYR-1 SVHRAAAPGAGYFVLVFAKGAFPAEMEGGPNEVTELELREAVSRYWIIDD
MAB_0961c|M.abscessus_ATCC_199 SISRAAAPDASYFVLVFAVGAFPPGIG--PNAVTEDELRAAVEPYWAIDE
MUL_0662|M.ulcerans_Agy99 SIVRAAAPGASYFVLVFDKAALSGDGP--INGVTEEELRDAVSKHWVIDE
MMAR_0089|M.marinum_M ----------------------------GPRELVRGERESFIGG------
TH_3824|M.thermoresistible__bu AIHRAAAPNARLFILTFAERPF--GDGHGPDGFTAEELRDTVGVLWTVDE
. * . :
Mb3725|M.bovis_AF2122/97 IRPAFIHVNPVTIPPQLAGAPVEFPPYDH-----DEKGRVK------FPA
Rv3699|M.tuberculosis_H37Rv IRPAFIHVNPVTIPPQLAGAPVEFPPYDH-----DEKGRVK------FPA
MAV_0404|M.avium_104 IRPSFIVSN----VPQIADAPFEFPAHER-----DERGRMK------MPA
MSMEG_6235|M.smegmatis_MC2_155 IRPAFIHAN----AVVFPDAP-ELPPHGY-----DDKGRIM------FPA
Mflv_1324|M.gilvum_PYR-GCK IRPALIHTN----VPKLPGMP-PPPLDML-----DDRGHLR------MQA
Mvan_5471|M.vanbaalenii_PYR-1 IRPALIDTN----VPKLPGMP-PPPLDML-----DDRGRLR------MQA
MAB_0961c|M.abscessus_ATCC_199 LSSAYIHSN---IPEVIPGANFEMPVFHK-----DEKGRNK------QPA
MUL_0662|M.ulcerans_Agy99 IRPARIHAN---LPPEVPGCPPSTLATNPRAASPSVRGCSQPIWADGTPH
MMAR_0089|M.marinum_M ------YRR------QHSGQAVGSP--AMNA---ADDG-----------C
TH_3824|M.thermoresistible__bu IRPAKLYVN------DFTGDLLDGPPTAYER---AADGKLM------FPG
. .. *
Mb3725|M.bovis_AF2122/97 YLLTAHKAG
Rv3699|M.tuberculosis_H37Rv YLLTAHKAG
MAV_0404|M.avium_104 YLLTAHKAQ
MSMEG_6235|M.smegmatis_MC2_155 FLLEAHRD-
Mflv_1324|M.gilvum_PYR-GCK FLLSAHKEP
Mvan_5471|M.vanbaalenii_PYR-1 FLLQAHKES
MAB_0961c|M.abscessus_ATCC_199 FLLQAHKR-
MUL_0662|M.ulcerans_Agy99 AVYSPANEG
MMAR_0089|M.marinum_M TRASSRR--
TH_3824|M.thermoresistible__bu FLVSAHKD-
. .