For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MRLEGAARSKVKAVLAVALVGLVVVGTGVALRNTFFRPTTITAYFPTATAIYPGDDVRVSGMKVGTIAAI RPDGTKAKMILHVDHNVPIPADAKAVIVAQNLVAARYVQLTPAYRSSGPVMADGAVIPIERTAVPVEWDE VKDQLMRLATELGPNSKVSSPSISRFIDSAADALGNGNGDKLRQTLAELSGVGRILANGSGNIVDIIKNL QTFIGALRDSNVQIVQFNDRLASLTSVLDDNKSDLDAALTDLSSAVGEVQRFIAGSRDATAEQITRLADL TQILVDNKMALKNVLHVTPNALANAYNDYDPDVGNVRGGVGIQNLTNPVFSICSQLGAMENVTSVESGKL CGLYLSPALKVFNPLMYFNFNYFPIPINPILAPAFDLQNVVYTEQRLAPGGEGPKPIAPELPPAVSAYTG LPGDTPEAPPPPPPPARIPGAAMPEPPPPSVPAEPSPPPANLSDMLLPAEGPQP*
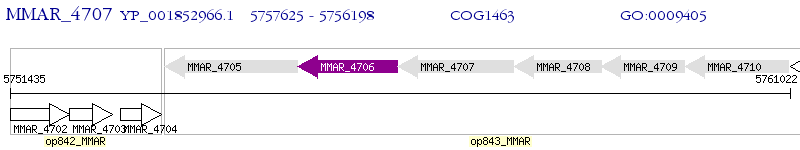
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_4707 | - | - | 100% (475) | MCE-family protein |
| M. marinum M | MMAR_4054 | mce3D_1 | 3e-73 | 36.23% (461) | MCE-family protein Mce3D_1 |
| M. marinum M | MMAR_4984 | mce4D | 2e-70 | 37.09% (488) | MCE-family protein Mce4D |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3526c | mce4D | 4e-67 | 36.57% (443) | MCE-family protein MCE4D |
| M. gilvum PYR-GCK | Mflv_2511 | - | 1e-147 | 56.39% (509) | virulence factor Mce family protein |
| M. tuberculosis H37Rv | Rv3496c | mce4D | 4e-67 | 36.57% (443) | MCE-family protein MCE4D |
| M. leprae Br4923 | MLBr_02592 | mce1D | 3e-67 | 33.27% (526) | putative secreted protein |
| M. abscessus ATCC 19977 | MAB_4150c | - | 5e-73 | 36.80% (462) | MCE-family protein |
| M. avium 104 | MAV_0951 | - | 0.0 | 70.17% (476) | virulence factor mce family protein |
| M. smegmatis MC2 155 | MSMEG_4787 | - | 1e-144 | 56.81% (470) | virulence factor mce family protein |
| M. thermoresistible (build 8) | TH_2817 | - | 1e-143 | 56.34% (481) | PUTATIVE virulence factor mce family protein |
| M. ulcerans Agy99 | MUL_3921 | mce3D_1 | 6e-72 | 35.65% (460) | MCE-family protein Mce3D_1 |
| M. vanbaalenii PYR-1 | Mvan_2854 | - | 1e-148 | 56.97% (495) | virulence factor Mce family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3526c|M.bovis_AF2122/97 ------MMGRVAMLTGSRGLRYATVIALVAALVGG-VYVLSSTGNKRTIV
Rv3496c|M.tuberculosis_H37Rv ------MMGRVAMLTGSRGLRYATVIALVAALVGG-VYVLSSTGNKRTIV
MAB_4150c|M.abscessus_ATCC_199 ------------MTTSKTWLKRGAFAMLSVALVGGLLYWVWPSRGTYKIV
MMAR_4707|M.marinum_M -------MMRLEGAARSKVKAVLAVALVGLVVVGTGVALRNTFFRPTTIT
MAV_0951|M.avium_104 --------MSPR-VPRNRLAALAAVVLVGLILAGTAVLVRNTFFGQKTIT
Mflv_2511|M.gilvum_PYR-GCK -----------MMATRKRWVAGTAVLLAILLVAGAVFLVRQVFFGPTTIT
MSMEG_4787|M.smegmatis_MC2_155 -------------MNRKRIAKGLAVGLVVLLVAGAAVLVRQVFLAPRTFQ
TH_2817|M.thermoresistible__bu --------MIATWRTAPRITRTLLIVLVALMLASSGYLLYQNRFGPITIS
Mvan_2854|M.vanbaalenii_PYR-1 --------MTVRGGSLRPARALLVVALTVLAVAGSGVLVYQHFFSPKTIV
MLBr_02592|M.leprae_Br4923 MSTIFDVRKLKLLRLSRAPVIVGSLVVVLALVVGFVGWRLYQKLTNNTVV
MUL_3921|M.ulcerans_Agy99 -------------MIRRTKTWLAVLLVGLLAAGVVVLLRTTEVVNRTNVV
. ..
Mb3526c|M.bovis_AF2122/97 GYFTSAVGLYPGDQVRVLGVPVGEIDMIEPRSSDVKITMSVSKDVKVPVD
Rv3496c|M.tuberculosis_H37Rv GYFTSAVGLYPGDQVRVLGVPVGEIDMIEPRSSDVKITMSVSKDVKVPVD
MAB_4150c|M.abscessus_ATCC_199 GHFASAVGLYPGDDVRILGVKAGRISAVEPREGDVKVTMEMPDKFKVPEG
MMAR_4707|M.marinum_M AYFPTATAIYPGDDVRVSGMKVGTIAAIRPDGTKAKMILHVDHNVPIPAD
MAV_0951|M.avium_104 AYFTTATAIYPNDEVRVSGVKVGNIKSIEPQGTQAKMTLKVDHDVPIPAD
Mflv_2511|M.gilvum_PYR-GCK AYFPTATAIYPGDEVRVSGVEVGTIDKIEPEGTQTKMTLKVDRGVQVPAN
MSMEG_4787|M.smegmatis_MC2_155 ALFTSATGIYPGDDVRVSGVKVGTIEAITPEGTQTRLTLKVDRGVPIPAD
TH_2817|M.thermoresistible__bu AYFRSATAVYPGDDVRVAGVRVGTIKSIEPMGTRAKVVLQVRRDVPIPAD
Mvan_2854|M.vanbaalenii_PYR-1 AYFRSATAIYAGDEVRVAGVRVGTIEAIEPQGTRAKMTLAVDRGVPIPAD
MLBr_02592|M.leprae_Br4923 VYFPAATALYTGDKVQIMGLRVGSIDSIVPAGDKMKVTFHYQNKYKVPAN
MUL_3921|M.ulcerans_Agy99 AYFENSNGVFTGDEVRILGVPVGKITSIEPQPESVRVSFWYDSKYIVPAD
* : .::..*.*:: *: .* * : * :: : :* .
Mb3526c|M.bovis_AF2122/97 VQAVIMSPNLVAARFIQLTPVYTG-----GAVLPDNGRIDLDRTAVPVEW
Rv3496c|M.tuberculosis_H37Rv VQAVIMSPNLVAARFIQLTPVYTG-----GAVLPDNGRIDLDRTAVPVEW
MAB_4150c|M.abscessus_ATCC_199 AQAIIMAPNLVTARFVQLTPAYTD-----GKALQDGASLGLDKTAVPIEW
MMAR_4707|M.marinum_M AKAVIVAQNLVAARYVQLTPAYRS----SGPVMADGAVIPIERTAVPVEW
MAV_0951|M.avium_104 AKAVIVASNLVSARYVQLSPAYRD----SGPVMPDGAVIPVERTAVPVEW
Mflv_2511|M.gilvum_PYR-GCK AKAVIVAQNLVAARYVQLTPAYRDG---DGATMADGGVIPKERTAIPVEW
MSMEG_4787|M.smegmatis_MC2_155 AKAVIVAQNLVAARYVQLAPAYRS----EGPTLPDDAVIGLDRTAVPVEW
TH_2817|M.thermoresistible__bu AEAIIVAQNLVSARYVQLTPPYGSAAAPDGPTMPDGGEIGLERTAVPIEW
Mvan_2854|M.vanbaalenii_PYR-1 AEAIIVAPNLVSARYVQLTPAWGASPETSGPTMADGGEIGQDRTAVPVEW
MLBr_02592|M.leprae_Br4923 ASAVILNPTLVASRSIQLEPPYKG-----GPALGDNAVIPLERTQVPVEW
MUL_3921|M.ulcerans_Agy99 AKAAILSPTLVTSRAIQLTPAYTG-----GPAMSDNAVIPRQRTAVPVEW
..* *: .**::* :** * : * .: *.. : ::* :*:**
Mb3526c|M.bovis_AF2122/97 DEVKEGLTRLAADLSPAAGELQGPLGAAINQAADTLD-GNGDSLHNALRE
Rv3496c|M.tuberculosis_H37Rv DEVKEGLTRLAADLSPAAGELQGPLGAAINQAADTLD-GNGDSLHNALRE
MAB_4150c|M.abscessus_ATCC_199 DEVKTELAKLSDSLGPQ-GQSKGPVSDFINQAATTFD-GNGDSFRNAVRE
MMAR_4707|M.marinum_M DEVKDQLMRLATELGPNSKVSSPSISRFIDSAADALGNGNGDKLRQTLAE
MAV_0951|M.avium_104 DEVKTQLMRLATDLGPKSGVSGTSVGRFIDSAANALD-GNGDKLRQTLAQ
Mflv_2511|M.gilvum_PYR-GCK DEVKTQLMRLSEELGPQAGASGTSISRFVDSAANAME-GNGDKLRETLRQ
MSMEG_4787|M.smegmatis_MC2_155 DEVKEQLTRLATELGPRSGVDGTSMARFIETTADAME-GNGEKLRNTISQ
TH_2817|M.thermoresistible__bu DEVKEQLTRLATELGPDGGVSDTALGRFIDSAADAMA-GNGGKLRQTISE
Mvan_2854|M.vanbaalenii_PYR-1 DEIKEQLTRLAAELGPASGVSGTSLGRFIDTTADAMA-GNGEKLRETIAQ
MLBr_02592|M.leprae_Br4923 DELRDSITKIISKLGPTKEQPKGPFGDVIESFADGLA-GKGKQINATLDS
MUL_3921|M.ulcerans_Agy99 DDVRAQLAKLTKELQPTEPGGVSPLGSVINTAADNLR-GEGANIRDTVIK
*::: : :: .* * ... :: * : *:* .:. :: .
Mb3526c|M.bovis_AF2122/97 LAQVAGRLGDSRGDIFGTVKNLQVLVDALSESDEQIVQFAGHVASVSQVL
Rv3496c|M.tuberculosis_H37Rv LAQVAGRLGDSRGDIFGTVKNLQVLVDALSESDEQIVQFAGHVASVSQVL
MAB_4150c|M.abscessus_ATCC_199 LSQAAGRLGDSRTDLFGTIKNLQTLVDALANSNEQIVQFSTHLASVSKVL
MMAR_4707|M.marinum_M LSGVGRILANGSGNIVDIIKNLQTFIGALRDSNVQIVQFNDRLASLTSVL
MAV_0951|M.avium_104 LSGVGRILANGSGNIVDIIKNLQTFVGALRDSNVQIVQFNDRLATLTSVV
Mflv_2511|M.gilvum_PYR-GCK LSGAARIFAEGSGNIVDIIKNLQIFVTALRDSKDQIVLFQNRLATLTSVV
MSMEG_4787|M.smegmatis_MC2_155 LSGVARILAEGSGNIVDIIKNLQTFVTALRDSNQQVVAFQNRLATLTSVV
TH_2817|M.thermoresistible__bu LSAVGRVLGEGSGNIVEILENLQTFVTALRDSNIQIVQFQDRLADVTSVV
Mvan_2854|M.vanbaalenii_PYR-1 LSGVGRILGEGSGDIVEVIQNLQIFVTALRDSNTQIVQFQDRLADVTSVV
MLBr_02592|M.leprae_Br4923 LSRALTALNEGRGDFFAVLRSLALFVNALHTDDQQFVALNQNLADFTTRL
MUL_3921|M.ulcerans_Agy99 LSQAFSALGDHSTDIFSTVKNLAILVSALQDSTNLMRQLNQNLATVTGLL
*: . : : ::. :..* :: ** . . : .:* .: :
Mb3526c|M.bovis_AF2122/97 ADSSANLDQTLGTLNQALSDIRGFLRENNSTLIETVNQLNDFAQTLS--D
Rv3496c|M.tuberculosis_H37Rv ADSSANLDQTLGTLNQALSDIRGFLRENNSTLIETVNQLNDFAQTLS--D
MAB_4150c|M.abscessus_ATCC_199 AASTENLGATLGALNQALTDVRGFLGDTNSALIDQVNKLSDFVKIFS--D
MMAR_4707|M.marinum_M DDNKSDLDAALTDLSSAVGEVQRFIAGSRDATAEQITRLADLTQILV--D
MAV_0951|M.avium_104 NDSKSDLDAALTDLSTAVGEVQRFIAGTRNQTSEQIARLADVTQILV--D
Mflv_2511|M.gilvum_PYR-GCK NDSRSDLDAALTDLSFAIGEVQRFVVGSRDATVEQIRSLGQVTQVLA--D
MSMEG_4787|M.smegmatis_MC2_155 DGSRSDLDAALKNLSVAVGEVQRFVAGSRNQTSEQIQRLADVTSNLV--D
TH_2817|M.thermoresistible__bu DGSRAELDAAIRDVADAVGVVERFIDGTRDQTAEQIQRLAALTRVLS--D
Mvan_2854|M.vanbaalenii_PYR-1 DGSRSDLDSAITTLSEAVGEVRRFVQGSRGQTVEQVQRLANVTQVLS--D
MLBr_02592|M.leprae_Br4923 AHSDTDLSSALQQFNSMLSTVQSFLVKNREVLAADVNNLATVNNTVVQPE
MUL_3921|M.ulcerans_Agy99 ANDPNEVANAVRNLGDTVGEVQRFVADNREALGTTSDKLAGVSQALN--D
. :: :: . : :. *: .. * . . :
Mb3526c|M.bovis_AF2122/97 QSENIEQVLHVAGPGITNFYNIYDPAQGTLNGLLSIPNFANPVQF-----
Rv3496c|M.tuberculosis_H37Rv QSENIEQVLHVAGPGITNFYNIYDPAQGTLNGLLSIPNFANPVQF-----
MAB_4150c|M.abscessus_ATCC_199 QSDDLEQILHVAPNGLANFYNIYNPAAGTLNGMLSIPNLANPVQF-----
MMAR_4707|M.marinum_M NKMALKNVLHVTPNALANAYNDYDPDVGNVRGGVGIQNLTNPVFS-----
MAV_0951|M.avium_104 HHMDLENILHVAPNALGNFFNDYNADTGTIVGGFGIMNFANPTFSGLMVP
Mflv_2511|M.gilvum_PYR-GCK NRLALENVLHITPNAIANFENIYYPNGGSVTGAFSLSNFSNPVWF-----
MSMEG_4787|M.smegmatis_MC2_155 QRLQVENLLHIAPNAFVNGYNIYNPDSGSPVGQFVFHNMSEPLHF-----
TH_2817|M.thermoresistible__bu NSMTVKNVLHAAPTALANGYNIYNPDTGGPRGSFAMNNFANPVTL-----
Mvan_2854|M.vanbaalenii_PYR-1 DSMVLKNILHAAPNALVNGYNIYNPDTGGPRGSFAMNNFANPVTM-----
MLBr_02592|M.leprae_Br4923 PLNGVETFLHVAPTAAANMNQLYHPAHGSVVGVPTITNFANPMQF-----
MUL_3921|M.ulcerans_Agy99 SLDDVKQFLHVAPNTLQNYVNIWQPAQGAVSAVPMLNNFANPISF-----
:: .** : * : : . * . : *:::*
Mb3526c|M.bovis_AF2122/97 ----ICGGSFDTAAGPSAPDYYRRAEICRERLGPVLRRLT------VNYP
Rv3496c|M.tuberculosis_H37Rv ----ICGGSFDTAAGPSAPDYYRRAEICRERLGPVLRRLT------VNYP
MAB_4150c|M.abscessus_ATCC_199 ----ICGGVFETGT---KTEYLKRAEICRERMGPVLRRAT------FNYA
MMAR_4707|M.marinum_M ----ICSQLGAMEN----VTSVESGKLCGLYLSPALKVFNPLMYFNFNYF
MAV_0951|M.avium_104 LPVPGCTAVGAIEN----ITAVESGKLCSLFLGPGLRVLN------FNNL
Mflv_2511|M.gilvum_PYR-GCK ----VCGLIGGIAN----TTAPETAKLCSQYLGPALRLVN------FGNL
MSMEG_4787|M.smegmatis_MC2_155 ----ICGAIGAVEN----VTAPETAKLCAQYLGPALRLLN------FNNV
TH_2817|M.thermoresistible__bu ----ICAAIGAVEN----VTAEASAKACAEYLGPALRLIN------FNYL
Mvan_2854|M.vanbaalenii_PYR-1 ----ICSAIAAVEN----VTAEESGKACAQYLGPALRLMN------FNYL
MLBr_02592|M.leprae_Br4923 ----ICSAIQAGSR----LGYQESAELCAQYLSPVLDAIK------FNYF
MUL_3921|M.ulcerans_Agy99 ----LCGAIQAASR----LGYEQSAKLCVQYLAPIIKNRQ------YNFL
* .: * :.* : .
Mb3526c|M.bovis_AF2122/97 PIMFHPLNTITAYKGQIIYDTP--------ATEAKSET---------PVP
Rv3496c|M.tuberculosis_H37Rv PIMFHPLNTITAYKGQIIYDTP--------ATEAKSET---------PVP
MAB_4150c|M.abscessus_ATCC_199 PILLHPINQISAYKGQIIYDTP--------ETEAKSQA---------PRE
MMAR_4707|M.marinum_M PIPINPILAPAFDLQNVVYTEQRLAPGGEGPKPIAPEL---------PPA
MAV_0951|M.avium_104 PIPINIFLQKSVDPSKILYSEPRLAPGGEGPKPGPPEI---------PPA
Mflv_2511|M.gilvum_PYR-GCK PFPINPYLRPAISPDRLIYTDPKLAPG-GAGPGDPPEI---------PPT
MSMEG_4787|M.smegmatis_MC2_155 PFPVSPYLMPSANPENLIYSEPNLAPGAGGGSPQPPET---------PPV
TH_2817|M.thermoresistible__bu PLPFNAYLGP--APQNVIYSEPGLAPGGGQRPPGPDEP---------PPA
Mvan_2854|M.vanbaalenii_PYR-1 PLPFNAYLGP--APQNVIYSEPGLAPGGGGTAPAPAET---------PPA
MLBr_02592|M.leprae_Br4923 PFGLNLFSTAEVLPKEVAYSEARLQPPSGYKDTTVPGIWVPDTPLSHRNT
MUL_3921|M.ulcerans_Agy99 PFGQNLFVGASARPNELTYSEDWLR-------------------------
*: .: *
Mb3526c|M.bovis_AF2122/97 ELTWVPAGGG--APVGN----------------------------PADLQ
Rv3496c|M.tuberculosis_H37Rv ELTWVPAGGG--APVGN----------------------------PADLQ
MAB_4150c|M.abscessus_ATCC_199 DLVWVRPNGAPPPPKPS----------------------------PQDLS
MMAR_4707|M.marinum_M VSAYTGLPGDTPEAP------------------------------PPPPP
MAV_0951|M.avium_104 VSAYTGLPGD-PVGP------------------------------PGAEP
Mflv_2511|M.gilvum_PYR-GCK VSAYTG-AGDIPPPPGWGAPPGPRGIYQPDPD------------APATPS
MSMEG_4787|M.smegmatis_MC2_155 LSAYN--PGPPPPPPFTGRPPG----------------------TPPPGA
TH_2817|M.thermoresistible__bu VSAYTG-AGDVAPPAGWHDPPHPPGAYTPDG-------------LPAIPQ
Mvan_2854|M.vanbaalenii_PYR-1 VSAYTG-VGDVPPPPGWSDPAHPPGAYAPHG-------------LPANPH
MLBr_02592|M.leprae_Br4923 QPGWIVAPGMQGQQVGPITAALMTPDSLAELMGGPDIEPIQSNLQTPPGP
MUL_3921|M.ulcerans_Agy99 -PDYIPPQAIPPQQA----------------------------------P
: .
Mb3526c|M.bovis_AF2122/97 SLLVPPAPGPAPA--------------PPAPGAG----------PGEHG-
Rv3496c|M.tuberculosis_H37Rv SLLVPPAPGPAPA--------------PPAPGAG----------PGEHG-
MAB_4150c|M.abscessus_ATCC_199 ALMLGPDEVNAPP--------------PPGTEVR----------PAEPGP
MMAR_4707|M.marinum_M PARIPGAAMPEPP--------------PPSVPAE--------PSPPP-AN
MAV_0951|M.avium_104 PARIPGAAMPLPP--------------PPSTPMP--------PPPPPEPG
Mflv_2511|M.gilvum_PYR-GCK PALYPGAPIPGPPNILSNIPPAPA-AGPAPTTVDGLLLPPGAPAAPP-VA
MSMEG_4787|M.smegmatis_MC2_155 AQLLPGAPPIVPPNVPSSVRDMLAPAGPPPGPV-----PETGPAPGP-LP
TH_2817|M.thermoresistible__bu PALFPGAPVPPGL--------------PAPAPSG----------PGP-AD
Mvan_2854|M.vanbaalenii_PYR-1 PALYPDAPIPPGV--------------PPPPPAR----------PAP-SS
MLBr_02592|M.leprae_Br4923 PTAYDEHPVPPPIGL--------QASVPIPSPRPDVVPGPVAPTPSPAAN
MUL_3921|M.ulcerans_Agy99 PAAAPPAQAPPPA-----------VGLPLPAEAP--------ATPDPAAG
. * .
Mb3526c|M.bovis_AF2122/97 -----------------GGG--
Rv3496c|M.tuberculosis_H37Rv -----------------GGG--
MAB_4150c|M.abscessus_ATCC_199 VPPGFPVTADDAPPPAIPGAPR
MMAR_4707|M.marinum_M LSDMLLPAEGPQP---------
MAV_0951|M.avium_104 VSGMLLPAQGPQQ---------
Mflv_2511|M.gilvum_PYR-GCK PNAPLLPAEGTPPS--------
MSMEG_4787|M.smegmatis_MC2_155 AEAPLEPAEGTPPA--------
TH_2817|M.thermoresistible__bu LQDLLLPAERHSDQ-QPGGTP-
Mvan_2854|M.vanbaalenii_PYR-1 IGDMLLPAEAAPAPPSTEGTP-
MLBr_02592|M.leprae_Br4923 VGDPPLPAELGAGQ--------
MUL_3921|M.ulcerans_Agy99 LRGIMVPQGVGS----------