For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MKQRLSSRGMRTVTIVALVAALVGGLYVLFSGGESGRKVVAYFTSAVGIYPGDEVRILGVPVGRIDSIEP RPADVKITMSVSNDVKIPRDAQALIVSPNLVAARFIQLTPVYTGGAVLPDGASIDLSRTAIPVEWDEVKE SLTQLAVQLSPAAGELQGPLGAAINQAADTLNGNGDSFHNALRELSQVAGRLGDSRNDIFSTVKNLQVLV DALSQSNEQIVQFAGHVASVSQVLADSSRNLDNTLGTLNQALSDIKGFLRENNSTLIETVNQLGDLTQTL SDQSDNIEQVLHVAGPGITNFYNIYDPAQGTLNGLLSIPNFANPVQFICGGSFDTAAGKSAPDYYRRAEL CRERLGPVLRRLTANYPPFLFHPLNTITAYKGQIIYDTPATQAKAETPVPELTWIPAHPAPQPAPGNTTD LQSLFVPQAPGPAPGSPPGYGPAPGPPPAPAPAGPDAGLSSPAPGAPLPAEQGAGG*
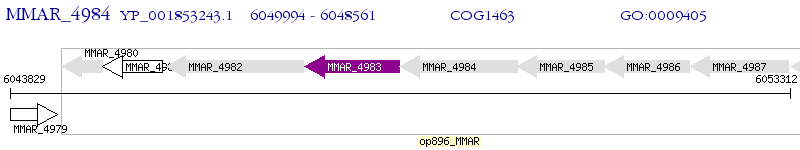
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_4984 | mce4D | - | 100% (477) | MCE-family protein Mce4D |
| M. marinum M | MMAR_4054 | mce3D_1 | 7e-80 | 38.33% (454) | MCE-family protein Mce3D_1 |
| M. marinum M | MMAR_0415 | mce1D | 9e-76 | 36.31% (537) | MCE-family protein Mce1D |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3526c | mce4D | 0.0 | 83.33% (438) | MCE-family protein MCE4D |
| M. gilvum PYR-GCK | Mflv_1559 | - | 1e-165 | 62.31% (467) | virulence factor Mce family protein |
| M. tuberculosis H37Rv | Rv3496c | mce4D | 0.0 | 83.33% (438) | MCE-family protein MCE4D |
| M. leprae Br4923 | MLBr_02592 | mce1D | 4e-70 | 35.21% (514) | putative secreted protein |
| M. abscessus ATCC 19977 | MAB_4150c | - | 1e-144 | 56.99% (458) | MCE-family protein |
| M. avium 104 | MAV_0660 | - | 0.0 | 75.42% (472) | virulence factor mce family protein |
| M. smegmatis MC2 155 | MSMEG_5897 | - | 1e-162 | 63.76% (447) | virulence factor mce family protein |
| M. thermoresistible (build 8) | TH_1934 | - | 1e-155 | 62.65% (423) | MCE-FAMILY PROTEIN MCE4D |
| M. ulcerans Agy99 | MUL_4059 | mce4D | 0.0 | 99.58% (477) | MCE-family protein Mce4D |
| M. vanbaalenii PYR-1 | Mvan_5199 | - | 1e-163 | 62.05% (469) | virulence factor Mce family protein |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4984|M.marinum_M -----MMK---QRLSSRGMRTVTIVALVAALVGG-LYVLFSGGESGRKVV
MUL_4059|M.ulcerans_Agy99 -----MMK---QRLSSRGMRTVTIVALVAALVGG-LYVLFSGGESGRKVV
Mb3526c|M.bovis_AF2122/97 -----MMGRVAMLTGSRGLRYATVIALVAALVGG-VYVLSSTGNK-RTIV
Rv3496c|M.tuberculosis_H37Rv -----MMGRVAMLTGSRGLRYATVIALVAALVGG-VYVLSSTGNK-RTIV
MAV_0660|M.avium_104 -----MMQR---LAGSRGLRYTTIIALVAVLVGG-VYVLTSQAKT-RKIV
Mflv_1559|M.gilvum_PYR-GCK -----MTQENSVT-RSRWLRPVLICVLVAVLAIG-AYLVWPSRTG-QKVT
Mvan_5199|M.vanbaalenii_PYR-1 -----------MK-RSRWLRTALVGVLVATLAIG-AYLVWPNRAG-HKVT
MSMEG_5897|M.smegmatis_MC2_155 -----MTHSNSTSNRGRWVRLALAALLLVTLGVG-IYQVWPSRGG-LKLT
TH_1934|M.thermoresistible__bu -----MTDTTTQR-NRRALRYLAVAILAIALTGG-ALQVWPKATS-RTLV
MAB_4150c|M.abscessus_ATCC_199 -----------MTTSKTWLKRGAFAMLSVALVGGLLYWVWPSRGT-YKIV
MLBr_02592|M.leprae_Br4923 MSTIFDVRKLKLLRLSRAPVIVGSLVVVLALVVGFVGWRLYQKLTNNTVV
: .* * .:.
MMAR_4984|M.marinum_M AYFTSAVGIYPGDEVRILGVPVGRIDSIEPRPADVKITMSVSNDVKIPRD
MUL_4059|M.ulcerans_Agy99 AYFTSAVGIYPGDEVRILGVPVGRIDSIEPRPADVKITMSVSNDVKIPRD
Mb3526c|M.bovis_AF2122/97 GYFTSAVGLYPGDQVRVLGVPVGEIDMIEPRSSDVKITMSVSKDVKVPVD
Rv3496c|M.tuberculosis_H37Rv GYFTSAVGLYPGDQVRVLGVPVGEIDMIEPRSSDVKITMSVSKDVKVPVD
MAV_0660|M.avium_104 GYFTSAVGLYPGDQVRVLGVPVGTIDTIDPRPTDVKITMSVSQDVKVPKD
Mflv_1559|M.gilvum_PYR-GCK AYFTSAVGLYPGDDVRVVGVPVGTIDAIDPRANDVKITMTIDSDVKLPAE
Mvan_5199|M.vanbaalenii_PYR-1 AYFTSAVGLYPGDDVRVVGVPVGTIDSIEPRAGDVKITMTIDGDVKVPAE
MSMEG_5897|M.smegmatis_MC2_155 AYFTNAVGLYPGDQVRVVGVPVGTIDSIEPRPSDVKIEMTLDRGVKVPAD
TH_1934|M.thermoresistible__bu AYFSSAVGIYPGDDVRVVGVPVGRIEAIEPQPDSVKITMRVNNDVQVPAD
MAB_4150c|M.abscessus_ATCC_199 GHFASAVGLYPGDDVRILGVKAGRISAVEPREGDVKVTMEMPDKFKVPEG
MLBr_02592|M.leprae_Br4923 VYFPAATALYTGDKVQIMGLRVGSIDSIVPAGDKMKVTFHYQNKYKVPAN
:*. *..:*.**.*:::*: .* *. : * .:*: : ::*
MMAR_4984|M.marinum_M AQALIVSPNLVAARFIQLTPVYTGGAVLPDGASIDLSRTAIPVEWDEVKE
MUL_4059|M.ulcerans_Agy99 AQALIVSPNLVAARFIQLTPVYTGGAVVPDGASIDLSRTAIPVEWDEVKE
Mb3526c|M.bovis_AF2122/97 VQAVIMSPNLVAARFIQLTPVYTGGAVLPDNGRIDLDRTAVPVEWDEVKE
Rv3496c|M.tuberculosis_H37Rv VQAVIMSPNLVAARFIQLTPVYTGGAVLPDNGRIDLDRTAVPVEWDEVKE
MAV_0660|M.avium_104 AKAIIMSPNLVAARFIQLTPAYTGGPVLADGASIGLDRTAVPVEWDEVKQ
Mflv_1559|M.gilvum_PYR-GCK AKALVISPNLVSARFIQLTPAYTGGAVLADGAEIGLDRTAVPVEWDEVKE
Mvan_5199|M.vanbaalenii_PYR-1 AKALVIAPNLVSARFIQLAPAYTGGTVMADGAEIGLDRTAVPVEWDEVKE
MSMEG_5897|M.smegmatis_MC2_155 AKALIIAPNLVAARFIQLTPAYTEGDEMADGTSIGLDRTAVPVEWDQVKE
TH_1934|M.thermoresistible__bu VRAVIMAPNLVSARFVQLTPAYTEGPTLEDGAVLDTDRTAVPVEWDQVKE
MAB_4150c|M.abscessus_ATCC_199 AQAIIMAPNLVTARFVQLTPAYTDGKALQDGASLGLDKTAVPIEWDEVKT
MLBr_02592|M.leprae_Br4923 ASAVILNPTLVASRSIQLEPPYKGGPALGDNAVIPLERTQVPVEWDELRD
. *::: *.**::* :** * *. * : *. : .:* :*:***:::
MMAR_4984|M.marinum_M SLTQLAVQLSPAAGELQGPLGAAINQAADTLNGNGDSFHNALRELSQVAG
MUL_4059|M.ulcerans_Agy99 SLTQLAVQLSPAAGELQGPLGAAINQAADTLNGNGDSFHNALRELSQVAG
Mb3526c|M.bovis_AF2122/97 GLTRLAADLSPAAGELQGPLGAAINQAADTLDGNGDSLHNALRELAQVAG
Rv3496c|M.tuberculosis_H37Rv GLTRLAADLSPAAGELQGPLGAAINQAADTLDGNGDSLHNALRELAQVAG
MAV_0660|M.avium_104 SLTQLAVQLGPTAGSMQGPLGAAINQAADTFDGKGESFHSALRELSQAAG
Mflv_1559|M.gilvum_PYR-GCK QLTQLSTQLGPNEDSVRGPLTDVVNQAADTFDGNGDTFRSAVRELSQTAG
Mvan_5199|M.vanbaalenii_PYR-1 QLTQLSTQLGPHEGSVQGPLTDVVNQAADTFDGNGDTFRRAVRELSQTAG
MSMEG_5897|M.smegmatis_MC2_155 QLTQLSSQLGPQEGSVQGPLAQVVNQAADTFDGNGDSFRNALRELSQTAG
TH_1934|M.thermoresistible__bu ELTKLSTQLGPGGQDVQGPLSAFVNQAADTFDGNGDSFRSALRELAQTAG
MAB_4150c|M.abscessus_ATCC_199 ELAKLSDSLGPQ-GQSKGPVSDFINQAATTFDGNGDSFRNAVRELSQAAG
MLBr_02592|M.leprae_Br4923 SITKIISKLGPTKEQPKGPFGDVIESFADGLAGKGKQINATLDSLSRALT
:::: .*.* . :**. ::. * : *:*. :. :: .*::.
MMAR_4984|M.marinum_M RLGDSRNDIFSTVKNLQVLVDALSQSNEQIVQFAGHVASVSQVLADSSRN
MUL_4059|M.ulcerans_Agy99 RLGDSRNDIFSTVKNLQVLVDALSQSNEQIVQFAGHVASVSQVLADSSRN
Mb3526c|M.bovis_AF2122/97 RLGDSRGDIFGTVKNLQVLVDALSESDEQIVQFAGHVASVSQVLADSSAN
Rv3496c|M.tuberculosis_H37Rv RLGDSRGDIFGTVKNLQVLVDALSESDEQIVQFAGHVASVSQVLADSSAN
MAV_0660|M.avium_104 RLGDSRGDIFGTVKNLQILVNALSSSNEQIVQFAGNVASVSQVLADSSRH
Mflv_1559|M.gilvum_PYR-GCK RLGDSRADLFGTIRNLQILVDALSRSNEQIVQFSNHVASVSQVLADATTD
Mvan_5199|M.vanbaalenii_PYR-1 RLGDSRTDLFGTIRNLQILVDALSNSNEQIVQFSNHVASVSQVLADATTD
MSMEG_5897|M.smegmatis_MC2_155 RLGDSRTDLFGTVRNLQILVDALSNSNEQIVQFTNHVASVSQVLADSANG
TH_1934|M.thermoresistible__bu RLGDSRTDLFGTVKNLQILIDALSNSNEQIVQFSGHLASVSQVLAESSTN
MAB_4150c|M.abscessus_ATCC_199 RLGDSRTDLFGTIKNLQTLVDALANSNEQIVQFSTHLASVSKVLAASTEN
MLBr_02592|M.leprae_Br4923 ALNEGRGDFFAVLRSLALFVNALHTDDQQFVALNQNLADFTTRLAHSDTD
*.:.* *:*..::.* :::** .::*:* : ::*..: ** :
MMAR_4984|M.marinum_M LDNTLGTLNQALSDIKGFLRENNSTLIETVNQLGDLTQTLSDQS--DNIE
MUL_4059|M.ulcerans_Agy99 LDNTLGTLNQALSDIKGFLRENNSTLIETVNQLGDLTQTLSDQS--DNIE
Mb3526c|M.bovis_AF2122/97 LDQTLGTLNQALSDIRGFLRENNSTLIETVNQLNDFAQTLSDQS--ENIE
Rv3496c|M.tuberculosis_H37Rv LDQTLGTLNQALSDIRGFLRENNSTLIETVNQLNDFAQTLSDQS--ENIE
MAV_0660|M.avium_104 LDTTLGTLNKALSDIRGFLHENNSTIVDTVNNLNDFAKTLSDQS--DNIE
Mflv_1559|M.gilvum_PYR-GCK LDITLGTLNQALTDVRGFLGESNEALIGQVNRLTDFTNLLTEHS--DDIE
Mvan_5199|M.vanbaalenii_PYR-1 LDVTLGTLNQALTDVRGFLGESNEALIGQVNKLTDFTNILTAQS--DDIE
MSMEG_5897|M.smegmatis_MC2_155 LDSTLDTLNQALSDVRGFLNESNDSLIAQVGKLADFTQILTDHS--EDIE
TH_1934|M.thermoresistible__bu LDGTLATLNQALNDVREFLDERNSTLIESVEKITDLSSLLTSHS--DDIE
MAB_4150c|M.abscessus_ATCC_199 LGATLGALNQALTDVRGFLGDTNSALIDQVNKLSDFVKIFSDQS--DDLE
MLBr_02592|M.leprae_Br4923 LSSALQQFNSMLSTVQSFLVKNREVLAADVNNLATVNNTVVQPEPLNGVE
*. :* :*. *. :: ** . .. : * .: . . . . :.:*
MMAR_4984|M.marinum_M QVLHVAGPGITNFYNIYDPAQGTLNGLLSIPNFANPVQFICGGSFDTAAG
MUL_4059|M.ulcerans_Agy99 QVLHVAGPGITNFYNIYDPAQGTLNGLLSIPNFANPVQFICGGSFDTAAG
Mb3526c|M.bovis_AF2122/97 QVLHVAGPGITNFYNIYDPAQGTLNGLLSIPNFANPVQFICGGSFDTAAG
Rv3496c|M.tuberculosis_H37Rv QVLHVAGPGITNFYNIYDPAQGTLNGLLSIPNFANPVQFICGGSFDTAAG
MAV_0660|M.avium_104 QVLHVAGPGIANFYNIYDPAQGTLNGLLSIPEFANPVQFICGGSFETAGG
Mflv_1559|M.gilvum_PYR-GCK QILHITPNGLSNFYNIYNPAQGTVGGLLTLPNFANPVQFICGGTFDAAAT
Mvan_5199|M.vanbaalenii_PYR-1 QILHITPNGLSNFYNIYNPAQGTVGGLLTLPNFGNPVQFICGGTFDAAAT
MSMEG_5897|M.smegmatis_MC2_155 QLLHVTPNGLANFYNIYNPAQGTVGGLLSLPNFANPVQFICGGTFDTGAN
TH_1934|M.thermoresistible__bu QLLHVAPNGLSNFYNIYNPAQGSMAGLLSLPNFANPVQFICAGSFESGAT
MAB_4150c|M.abscessus_ATCC_199 QILHVAPNGLANFYNIYNPAAGTLNGMLSIPNLANPVQFICGGVFETGTK
MLBr_02592|M.leprae_Br4923 TFLHVAPTAAANMNQLYHPAHGSVVGVPTITNFANPMQFICS-AIQAGSR
.**:: . :*: ::*.** *:: *: ::.::.**:****. :::.
MMAR_4984|M.marinum_M KSAPDYYRRAELCRERLGPVLRRLTANYPPFLFHPLNTITAYKGQIIYDT
MUL_4059|M.ulcerans_Agy99 KSAPDYYRRAELCRERLGPVLRRLTANYPPFLFHPLNTITAYKGQIIYDT
Mb3526c|M.bovis_AF2122/97 PSAPDYYRRAEICRERLGPVLRRLTVNYPPIMFHPLNTITAYKGQIIYDT
Rv3496c|M.tuberculosis_H37Rv PSAPDYYRRAEICRERLGPVLRRLTVNYPPIMFHPLNTITAYKGQIIYDT
MAV_0660|M.avium_104 PRAPDYYKRAELCRERLGPVLRRLTVNYPPLMFHPLNTITAYKGQIIYDT
Mflv_1559|M.gilvum_PYR-GCK ---PDNYKRAEICRQRMGPVFKRITMNYPPLLFHPINSITAYKGQIIYDT
Mvan_5199|M.vanbaalenii_PYR-1 ---TDNYKRAEICRQRMGPVFKRIAMNYPPLLFHPINSITAYKGQIIYDT
MSMEG_5897|M.smegmatis_MC2_155 ---PENYKRAEICRQRMGPVLKRITMNYPPVLFHPINSITAYKGQIIYDT
TH_1934|M.thermoresistible__bu ---TDNYKRTEICRQRMAPVLRRIMMNYPPMMFHPINSITAYKGQIIYDT
MAB_4150c|M.abscessus_ATCC_199 ---TEYLKRAEICRERMGPVLRRATFNYAPILLHPINQISAYKGQIIYDT
MLBr_02592|M.leprae_Br4923 ---LGYQESAELCAQYLSPVLDAIKFNYFPFGLNLFSTAEVLPKEVAYSE
. :*:* : :.**: ** *. :: :. . :: *.
MMAR_4984|M.marinum_M PATQAKA---ETPVP--------------ELTWIPAHPAPQ-------PA
MUL_4059|M.ulcerans_Agy99 PATQAKA---ETPVP--------------ELTWIPAHPAPQ-------PA
Mb3526c|M.bovis_AF2122/97 PATEAKS---ETPVP--------------ELTWVPAGGG---------AP
Rv3496c|M.tuberculosis_H37Rv PATEAKS---ETPVP--------------ELTWVPAGGG---------AP
MAV_0660|M.avium_104 PETQAKS---ATPVP--------------QLTWIPAKGAQTP------PA
Mflv_1559|M.gilvum_PYR-GCK PATEAKA---ETPVP--------------YLQWQNAPGVTP-------PK
Mvan_5199|M.vanbaalenii_PYR-1 PATEAKA---ETPVP--------------YLQWQNAPGVTP-------PQ
MSMEG_5897|M.smegmatis_MC2_155 PETQAKA---ETPVP--------------YLQWQNAPGVTP-------PQ
TH_1934|M.thermoresistible__bu PATEAKA---RTPIP--------------QLQWQPNPGANP-------PD
MAB_4150c|M.abscessus_ATCC_199 PETEAKS---QAPRE--------------DLVWVRPNGAPP-------PP
MLBr_02592|M.leprae_Br4923 ARLQPPSGYKDTTVPGIWVPDTPLSHRNTQPGWIVAPGMQGQQVGPITAA
. :. : :. * .
MMAR_4984|M.marinum_M PGNTTDLQSLFVPQAP---------------------GPAPGSPPGYGPA
MUL_4059|M.ulcerans_Agy99 PGNTTDLQSLFVPQAP---------------------GPTPGSPPGYGPA
Mb3526c|M.bovis_AF2122/97 VGNPADLQSLLVPPAP---------------------GPAP---------
Rv3496c|M.tuberculosis_H37Rv VGNPADLQSLLVPPAP---------------------GPAP---------
MAV_0660|M.avium_104 VQNPADLQALLVPTAPQ-------------------SGPAPAGG-----A
Mflv_1559|M.gilvum_PYR-GCK VAPDADLTSLLVPAEG----------------------AEASTSHAGGPE
Mvan_5199|M.vanbaalenii_PYR-1 IAPDADLTSLLVPAEG----------------------PEASTSHAGGPE
MSMEG_5897|M.smegmatis_MC2_155 IPADANLSDFIVPPD-----------------------PATATEHGG---
TH_1934|M.thermoresistible__bu LPPDTDLSELILPPA-----------------------------------
MAB_4150c|M.abscessus_ATCC_199 KPSPQDLSALMLGPDEV-------------------NAPPPPGTEVRPAE
MLBr_02592|M.leprae_Br4923 LMTPDSLAELMGGPDIEPIQSNLQTPPGPPTAYDEHPVPPPIGLQASVPI
.* ::
MMAR_4984|M.marinum_M PGPPPAPAP--AGPDAGLSSPAPGAPLPAEQGAGG
MUL_4059|M.ulcerans_Agy99 PGPPPAPAP--AGPDAGLSSPAPGAPLPAEQGAGG
Mb3526c|M.bovis_AF2122/97 --APPAPG---AG--------------PGEHGGGG
Rv3496c|M.tuberculosis_H37Rv --APPAPG---AG--------------PGEHGGGG
MAV_0660|M.avium_104 PAPGPAPGSA-FGPRPGPPPGQAPGGLGADLGGGG
Mflv_1559|M.gilvum_PYR-GCK TGPAPAAPA-----------------APGPAEAGR
Mvan_5199|M.vanbaalenii_PYR-1 TGPAPAAPV-----------------APGPAEAGR
MSMEG_5897|M.smegmatis_MC2_155 --PAAAPGP-----------------APAPGGAGG
TH_1934|M.thermoresistible__bu ---------------------------PEESEAGG
MAB_4150c|M.abscessus_ATCC_199 PGPVPPGFP----------VTADDAPPPAIPGAPR
MLBr_02592|M.leprae_Br4923 PSPRPDVVPGPVAPTPSPAANVGDPPLPAELGAGQ
.