For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MATTGYDVIVIGLGGMGSAAAYHLAARGRRVLGLERHQPAHDKGSSHGGSRIIRQSYFEDPGYVPLLLRS YELWERLAIDAQSDIYRLTGGLFIGPPDCATVAGSLRASREWDLPHEVLDAPQIAARFPNFTPQPTDIAL YEAKAGLARPELTVRAHLELAKRSGATLQFEEPVLNWTTTASGVRVTTGRDTYTAEQLVVCPGAWAPELL SQLGIPITVQRQVMYWLDPVGGTAGFRDHPIFIAENDSGAQIYGFPAIDGPRGGVKVAFFRKGIACTPDT IDRVVGPQEIHEMITRVGELLPALAGTCLQAATCMYSNTPDQHFVIARHPYCPAVTVACGFSGHGFKFVP VVGEILADLVTEGATSQPISLFDPQRLVTV
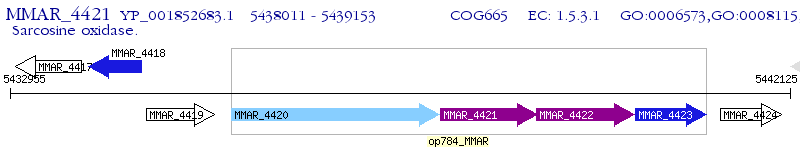
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_4421 | solA | - | 100% (380) | dehydrogenase |
| M. marinum M | MMAR_2926 | - | 2e-11 | 30.84% (214) | glycine/D-amino acid oxidase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3081 | solA | 1e-171 | 75.67% (374) | N-methyltryptophan oxidase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. avium 104 | MAV_0142 | - | 2e-06 | 27.27% (220) | FAD dependent oxidoreductase, putative |
| M. smegmatis MC2 155 | MSMEG_6264 | - | 4e-20 | 27.51% (378) | putative oxidoreductase |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_5513 | - | 3e-18 | 26.95% (371) | FAD dependent oxidoreductase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4421|M.marinum_M MATTGYDVIVIGLGGMGSAAAYHLAARGRRVLGLERHQPAHDKGSSHGGS
Mflv_3081|M.gilvum_PYR-GCK ---MTYDVIVIGLGGMGSAAAYHLAARGQRVLGLEKFTPAHDRGSSHGGS
MSMEG_6264|M.smegmatis_MC2_155 -MSITADVVIVGGGLEGTAAAWALSQRGVTDVVVAERNTVG--SGMTGKS
Mvan_5513|M.vanbaalenii_PYR-1 -MTETADVVIVGGGLEGAAAAWALSRRGVTDVVVAERNTVG--SGMTGKS
MAV_0142|M.avium_104 ------MIVVIGAGVCGLAAAHELSRRGHDDVVVLEKGQPF--GEQSAGL
::::* * * *** *: ** : : . . .
MMAR_4421|M.marinum_M RIIRQSYFEDPGYVPLLLRSYELWERLAIDAQSDI-YRLTGGLF-IGPPD
Mflv_3081|M.gilvum_PYR-GCK RIIRQSYFEDPAYVPLLLRAYELWAGLAADSGRDV-YRMTGGLF-LGPPD
MSMEG_6264|M.smegmatis_MC2_155 SGIVRCHYGVSSLAAMAAVGLDVFEKAEEIFGDDIGFRQTGYVVGVGEQN
Mvan_5513|M.vanbaalenii_PYR-1 SGIVRCHYGVSSLAAMAADGLEVFENGEDIFGTDIGFRQTGYVVGVGEQN
MAV_0142|M.avium_104 ARIFRIAHQREALCRLAFAARAGWQRWEAEFGVGRLLGSEGFIA-----A
* : . . : . : . * :
MMAR_4421|M.marinum_M CATVAGSLRASREWDLPHEVLDAPQIAARFPNFTPQPTDIALYEAKAGLA
Mflv_3081|M.gilvum_PYR-GCK CLTVAGSLRASREWGLPHDLLDGKEVRARFPNFTPHPGDVGLYEANAGFA
MSMEG_6264|M.smegmatis_MC2_155 VDALRKSLAAQRQVGVQTEEIDASEVAKLWPWADLEPFAAFGWEARGGYG
Mvan_5513|M.vanbaalenii_PYR-1 VESLRKSLAAQRAVGVDTEEIDKDEVAKLWPFADLTPFAAFGWEARGGYG
MAV_0142|M.avium_104 GAAADGVAQAMKRAGAAFSRLDRDGIAERIP-FADVPWETGIFDPLGG--
: * : . . :* : * * ::. .*
MMAR_4421|M.marinum_M RPELTVRAHLELAKRSGATLQFEEPVLNWTTTASGVR--VTTGRDTYTAE
Mflv_3081|M.gilvum_PYR-GCK RPELTVRAHLDLAEKAGATLRFGADVLDWSESSGGVT--VRTPGETFTAG
MSMEG_6264|M.smegmatis_MC2_155 DAYQTAQAFAIAARAAGVRIRQGATVTELLMGADRVTGVRLADGTEVSAG
Mvan_5513|M.vanbaalenii_PYR-1 DAYQTAQAFSTAARAAGARVRQGTAVTDLLVDGDRVTGVRTADGSEIFAG
MAV_0142|M.avium_104 -SLRIRRALTALARR--VVIRCGEVVS---VADDGRT--VLADGTVLRAD
. :* *. . :: * . : *
MMAR_4421|M.marinum_M QLVVCPGAWAPELLSQLG--IPITVQRQVMYWLDP---VGGTAGFRDHP-
Mflv_3081|M.gilvum_PYR-GCK QLVICPGAWAPQLLAEFG--IPITVERQVLYWLDP---VGGTASFVDHP-
MSMEG_6264|M.smegmatis_MC2_155 TVVVATGAWTRPFLAPYGVDIPIRVIREQIVTISPGLDIGAVPVFSDLVS
Mvan_5513|M.vanbaalenii_PYR-1 TVVVATGVWTRPFLARYGIDVPIRVIREQIVLIDPGVEVGKVPVFSDLVS
MAV_0142|M.avium_104 RVLICAGVATPKLFGPLG------------------------VGFVPHTR
:::..*. : ::. * *
MMAR_4421|M.marinum_M -IFIAENDSGAQIYGFPAIDGPRGGVKVAFFRKGIACTPDTIDRVVGPQE
Mflv_3081|M.gilvum_PYR-GCK -IFIDENASGAQIYGFPAIDGPRGGVKVAFFRKGVECTPETIDRTVHDHE
MSMEG_6264|M.smegmatis_MC2_155 LQYVRPELGGEILFGNSDLG------------HGESADPDNYLNRATEEF
Mvan_5513|M.vanbaalenii_PYR-1 LQYVRPEVGGEILFGNSDLPG---------YDAIDPADPDDYLNRATDGF
MAV_0142|M.avium_104 FTYRGADATGAACLSAPEGYG-------------LPLGGTGRWAFGQEVA
: : * . .
MMAR_4421|M.marinum_M IHEMITRVGELLPALAGTCLQAATC-MYSNTPDQHFVIARHPYCPAVTVA
Mflv_3081|M.gilvum_PYR-GCK VREMRDRVAELLPALDGPCVHSATC-MYANTPDQHFVIARHPDSERVTVA
MSMEG_6264|M.smegmatis_MC2_155 VDITVEKVGTRFPGLTDASITGSYAGCYDVTPDWNPVISETD-VDGLIVA
Mvan_5513|M.vanbaalenii_PYR-1 VDLTVDKVGTRFPGFPDAAISGSYAGCYDVTPDWNPIISVAP-LDGLVIA
MAV_0142|M.avium_104 DPATVRALFPSLVAVDRVDCVSVRAPWLDSGGDGWTVARRGR------VL
: : .. . . * : :
MMAR_4421|M.marinum_M CGFSGHGFKFVPVVGEILADLVTEGATSQP--------ISLFDPQRLVTV
Mflv_3081|M.gilvum_PYR-GCK CGFSGHGFKFVPVVGEILADLAISGATAHP--------ISLFDPRRLVTT
MSMEG_6264|M.smegmatis_MC2_155 AGFSGHGFKIAPAVGRLVADLVVDGHSSDPRIPETDFRLSRFAEDNLLKS
Mvan_5513|M.vanbaalenii_PYR-1 AGFSGHGFKIAPAVGRLVADIVVDGHSSDSRIPASDFRFSRFAEDDPLKT
MAV_0142|M.avium_104 AFVGANLMKFGPLLGELLARAALS---------------DDIPAELLLD-
. ...: :*: * :*.::* . . . : :
MMAR_4421|M.marinum_M -----------
Mflv_3081|M.gilvum_PYR-GCK R----------
MSMEG_6264|M.smegmatis_MC2_155 PYPYVGAGEMR
Mvan_5513|M.vanbaalenii_PYR-1 PYPYVGAGQMR
MAV_0142|M.avium_104 -----------