For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VAGVAVKFPPNRYQQAEAIGALTEFAGPEFRRFALSSGVEARHTALPLERYAELSGFTEANDAFVDVALD LGEQALRAALDEAKVRPSEVDVIFSTTVTGLAVPTLEARLAHRVGLREDVKRIPLFGLGCVAGAAGVARM HDYLRGFPDQVGALLAVELCSLTIQRHDNSVANLVASSLFGDAAAAVVTKGAHRVRGAGAASNGYAGPRV LATRSRIYPNTENVMGWKIGTSGFQIVLSVDVATVTEKYLGEDVRSFLADHGLTPEDVTTWVCHPGGPRV IEAVETVLGLPEDALAHTRNSLRNNGNLSSVSILDVLRANLADPPPAGSIGLMIAMGPAFCSELVLLTW
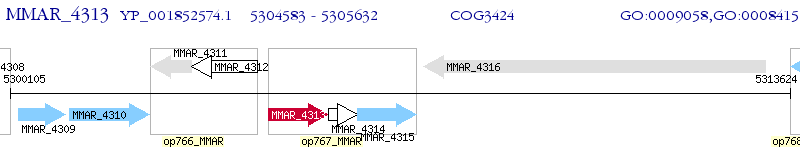
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_4313 | - | - | 100% (349) | chalcone synthase |
| M. marinum M | MMAR_2470 | pks10 | e-101 | 53.67% (354) | chalcone synthase, Pks10 |
| M. marinum M | MMAR_2474 | pks11_1 | 6e-99 | 51.41% (354) | chalcone synthase, Pks11_1 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1688 | pks10 | 1e-102 | 54.24% (354) | chalcone synthase |
| M. gilvum PYR-GCK | Mflv_5140 | - | 1e-100 | 52.81% (356) | chalcone/stilbene synthase domain-containing protein |
| M. tuberculosis H37Rv | Rv1660 | pks10 | 1e-102 | 54.24% (354) | chalcone synthase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. avium 104 | MAV_1280 | - | 1e-157 | 80.52% (349) | transferase |
| M. smegmatis MC2 155 | MSMEG_0808 | - | 2e-98 | 53.95% (354) | chalcone synthase Pks10 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_1656 | pks11 | 4e-98 | 50.85% (354) | chalcone synthase, Pks11 |
| M. vanbaalenii PYR-1 | Mvan_1196 | - | 1e-100 | 53.39% (354) | chalcone/stilbene synthase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1688|M.bovis_AF2122/97 ----------MSVIAGVFGALPPYRYSQRELTDSFVSIPDFEGYEDIVRQ
Rv1660|M.tuberculosis_H37Rv ----------MSVIAGVFGALPPYRYSQRELTDSFVSIPDFEGYEDIVRQ
MUL_1656|M.ulcerans_Agy99 ----------MSVIAGVFGALPPHRYSQSEITESFLEFPGLQEHEEIVRR
Mflv_5140|M.gilvum_PYR-GCK ---MTDVHTPVSVIAGVQGALPPHRYSQAEVTDAFLAAPAFAEVGDLLRS
Mvan_1196|M.vanbaalenii_PYR-1 ------MNDRISVIAGVQGALPPYRYSQAEITERFVAVPAFAELEDVVRG
MSMEG_0808|M.smegmatis_MC2_155 MENTAFTPSGTCTIAGVHGVLPPHRYTQAEVTEALLDLPGYADFEGIVRS
MMAR_4313|M.marinum_M -------------MAGVAVKFPPNRYQQAEAIGALTEFAG-----PEFRR
MAV_1280|M.avium_104 -------MGESPSITGVAVKFPPNRYTQSEAIRALTDIAG-----PEFRR
::** :** ** * * : . .*
Mb1688|M.bovis_AF2122/97 LHASAKVNSRHLVLPLEKYPKLTDFGEANKIFIEKAVDLGVQALAGALDE
Rv1660|M.tuberculosis_H37Rv LHASAKVNSRHLVLPLEKYPKLTDFGEANKIFIEKAVDLGVQALAGALDE
MUL_1656|M.ulcerans_Agy99 MHAAAKVNSRQFVLPIEQYPALTDFGDTNEIFIEKAVDLGIEALMGALDE
Mflv_5140|M.gilvum_PYR-GCK LHTNAKVDARHLVLPLEQYPDLLDFGDANDLYIHHAVELGCQALTAALDE
Mvan_1196|M.vanbaalenii_PYR-1 LHKHAKVDARHFVLPMDRYPTLVDFGETNDLYLEHSLDLACEALTAALDE
MSMEG_0808|M.smegmatis_MC2_155 LHKSAKVNSRYLTLPLEEYAALDDFGETNGLFIEHATELGCAALTGALDE
MMAR_4313|M.marinum_M FALSSGVEARHTALPLERYAELSGFTEANDAFVDVALDLGEQALRAALDE
MAV_1280|M.avium_104 FAHSSGVEFRNTALRLPRYRELSGFSEANDAYLEVALDLGEQALVAALDR
: : *: * .* : .* * .* ::* ::. : :*. ** .***.
Mb1688|M.bovis_AF2122/97 SGLRPEDLDVLITATVTGLAVPSLDARIAGRLGLRADVRRVPLFGLGCVA
Rv1660|M.tuberculosis_H37Rv SGLRPEDLDVLITATVTGLAVPSLDARIAGRLGLRADVRRVPLFGLGCVA
MUL_1656|M.ulcerans_Agy99 AGLTPQDIDLIITTTVTGVAVPSLDARIAGRLGLRPDVRRMPLFGLGCVA
Mflv_5140|M.gilvum_PYR-GCK AGLAPSDLDVIMSTTVTGIAVPSLDARIAAKLGLRPDVRRVPLFGLGCVA
Mvan_1196|M.vanbaalenii_PYR-1 AGLGPRDLDMIMTTTVTGIAVPSLDARIADRLGLRPDVRRVPLFGLGCVA
MSMEG_0808|M.smegmatis_MC2_155 AGLRPEDVDLIITTTVTGAVVPSLDARIAGRVGLRPDVRRVPIFGLGCVA
MMAR_4313|M.marinum_M AKVRPSEVDVIFSTTVTGLAVPTLEARLAHRVGLREDVKRIPLFGLGCVA
MAV_1280|M.avium_104 AKVKPSEVDIVFSTTVTGLAVPTLEARLATRVGLRPDVKRVPLFGLGCVA
: : * ::*:::::**** .**:*:**:* ::*** **:*:*:*******
Mb1688|M.bovis_AF2122/97 GAAGVARLHDYLRGAPDGVAALVSVELCSLTYPGYKPTLPGLVGSALFAD
Rv1660|M.tuberculosis_H37Rv GAAGVARLHDYLRGAPDGVAALVSVELCSLTYPGYKPTLPGLVGSALFAD
MUL_1656|M.ulcerans_Agy99 GAAGVARLHDYLRGAPDGVAVLIAVELCSLTYPAVKPTVSGIVGTALFGD
Mflv_5140|M.gilvum_PYR-GCK GAAGIARMHDYLRGAPGHVAALISVELCSLTRPALAPTIGSLVGSALFGD
Mvan_1196|M.vanbaalenii_PYR-1 GAAGIARMHDYLRGAPDHIAALVSVELCSLTYPAVEPTMASLVGSALFGD
MSMEG_0808|M.smegmatis_MC2_155 GAAGVARLNDYLRGAPDKVAVLVSVELCSLTH-KHTPSMPTLVAGALFGD
MMAR_4313|M.marinum_M GAAGVARMHDYLRGFPDQVGALLAVELCSLTIQRHDNSVANLVASSLFGD
MAV_1280|M.avium_104 GAAGVARMHDYLRAFPDQTAALLAVELCSLTIQRHDTSIANLVATSLFGD
****:**::****. *. ..*::******* :: :*. :**.*
Mb1688|M.bovis_AF2122/97 GAAAVVAAGVKR-----AQDIGADGPDILDSRSHLYPDSLRTMGYDVGSA
Rv1660|M.tuberculosis_H37Rv GAAAVVAAGVKR-----AQDIGADGPDILDSRSHLYPDSLRTMGYDVGSA
MUL_1656|M.ulcerans_Agy99 GAAAVVAVGDRR-----AEQIRAGGPDILDSRSRLYPESLDIMGWDVGSH
Mflv_5140|M.gilvum_PYR-GCK GAAAVVAVGEER-----AGHIRASGPDVLDSRSRMYPDSLDIMGWSVNHN
Mvan_1196|M.vanbaalenii_PYR-1 GAAAVIAVGAER-----AETIGATGPEVLDTRSRMYPDSLDTMGWRVGAS
MSMEG_0808|M.smegmatis_MC2_155 GASAVVAVGAGR-----AEQLDPIGPDVLDSRSHLYPDSLHAMGWDIGTK
MMAR_4313|M.marinum_M AAAAVVTKGAHRVRGAGAASNGYAGPRVLATRSRIYPNTENVMGWKIGTS
MAV_1280|M.avium_104 GAAAVITEGARR---AGAEHT---GPRILATRSRIYPDTEEVMGWKIGGD
.*:**:: * * * ** :* :**::**:: **: :.
Mb1688|M.bovis_AF2122/97 GFELVLSRDLAAVVEQYLGNDVTTFLASHGLSTTDVGAWVTHPGGPKIIN
Rv1660|M.tuberculosis_H37Rv GFELVLSRDLAAVVEQYLGNDVTTFLASHGLSTTDVGAWVTHPGGPKIIN
MUL_1656|M.ulcerans_Agy99 GLQLRLAPELTNLIEEYLPDDVDAFLAMHDLTKNDIGAWVSHPGGPKVLD
Mflv_5140|M.gilvum_PYR-GCK GFQLVLSPELPALIERYLPDDMASFLGEHDLGIGDIATWVSHPGGPKVLE
Mvan_1196|M.vanbaalenii_PYR-1 GFQLILSPNLPELIERYLADDMAGFLGAHDLQIGDIAAWVSHPGGPKVME
MSMEG_0808|M.smegmatis_MC2_155 GFEIVLSAEVPGFVGRYLGDDVTEFLGEHGVATDDVSAWVSHPGGPKVIE
MMAR_4313|M.marinum_M GFQIVLSVDVATVTEKYLGEDVRSFLADHGLTPEDVTTWVCHPGGPRVIE
MAV_1280|M.avium_104 GFRIVLSADVANIAEKYLGEDVRDFLADHGLAPRDVTTWVCHPGGPRVIE
*:.: *: ::. . .** :*: **. *.: *: :** *****::::
Mb1688|M.bovis_AF2122/97 AITETLDLSPQALELTWRSLGEIGNLSSASVLHVLRDTIAKPPPSGSPGL
Rv1660|M.tuberculosis_H37Rv AITETLDLSPQALELTWRSLGEIGNLSSASVLHVLRDTIAKPPPSGSPGL
MUL_1656|M.ulcerans_Agy99 AIANSLGLPSDALELTWRSLGEIGNLSSASVLRILRDTIAKVPPSGSPGL
Mflv_5140|M.gilvum_PYR-GCK AIASALDLPDDALELSWRSLAEIGNLSSSSVLHILRDTIAKKPPAATPGL
Mvan_1196|M.vanbaalenii_PYR-1 AISETLGLPDDALDLAWRSLAEVGNLSSSSVLHVLRDTRAKKPPAGTPGL
MSMEG_0808|M.smegmatis_MC2_155 AIIETLDLPSDALDLTWQSLAEVGNLSSSSVLHVLRDTIRKRPPAGTPGV
MMAR_4313|M.marinum_M AVETVLGLPEDALAHTRNSLRNNGNLSSVSILDVLRANLADPPPAGSIGL
MAV_1280|M.avium_104 AVESVLDLPADALDHTRNSLRENGNLSSVSVLDVLAANLADPPAPGSIGL
*: *.*. :** : .** : ***** *:* :* . . *...: *:
Mb1688|M.bovis_AF2122/97 MIAMGPGFCSELVLLRWH
Rv1660|M.tuberculosis_H37Rv MIAMGPGFCSELVLLRWH
MUL_1656|M.ulcerans_Agy99 MLAMGPGFCAELVLLRWH
Mflv_5140|M.gilvum_PYR-GCK LMAMGPGFCSELVLLRWH
Mvan_1196|M.vanbaalenii_PYR-1 LMAMGPGFCSELVLLRWR
MSMEG_0808|M.smegmatis_MC2_155 LMAMGPGFCSELVLLRWR
MMAR_4313|M.marinum_M MIAMGPAFCSELVLLTW-
MAV_1280|M.avium_104 MIAMGPAFCSELVLLAF-
::****.**:***** :