For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NDRISVIAGVQGALPPYRYSQAEITERFVAVPAFAELEDVVRGLHKHAKVDARHFVLPMDRYPTLVDFGE TNDLYLEHSLDLACEALTAALDEAGLGPRDLDMIMTTTVTGIAVPSLDARIADRLGLRPDVRRVPLFGLG CVAGAAGIARMHDYLRGAPDHIAALVSVELCSLTYPAVEPTMASLVGSALFGDGAAAVIAVGAERAETIG ATGPEVLDTRSRMYPDSLDTMGWRVGASGFQLILSPNLPELIERYLADDMAGFLGAHDLQIGDIAAWVSH PGGPKVMEAISETLGLPDDALDLAWRSLAEVGNLSSSSVLHVLRDTRAKKPPAGTPGLLMAMGPGFCSEL VLLRWR*
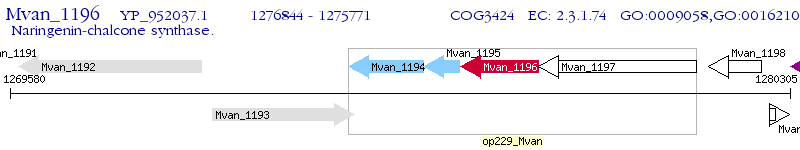
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_1196 | - | - | 100% (357) | chalcone/stilbene synthase domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1688 | pks10 | 1e-141 | 66.76% (352) | chalcone synthase |
| M. gilvum PYR-GCK | Mflv_5140 | - | 1e-164 | 78.69% (352) | chalcone/stilbene synthase domain-containing protein |
| M. tuberculosis H37Rv | Rv1660 | pks10 | 1e-141 | 66.76% (352) | chalcone synthase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1141c | - | 2e-06 | 26.04% (288) | 3-oxoacyl-(acyl carrier protein) synthase III |
| M. marinum M | MMAR_2474 | pks11_1 | 1e-145 | 68.47% (352) | chalcone synthase, Pks11_1 |
| M. avium 104 | MAV_3110 | - | 1e-142 | 68.18% (352) | Pks10 protein |
| M. smegmatis MC2 155 | MSMEG_0808 | - | 1e-140 | 67.43% (350) | chalcone synthase Pks10 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_1656 | pks11 | 1e-145 | 68.47% (352) | chalcone synthase, Pks11 |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_1196|M.vanbaalenii_PYR-1 ------MNDRISVIAGVQGALPPYRYSQAEITERFVAVPAFAELEDVVRG
Mflv_5140|M.gilvum_PYR-GCK ---MTDVHTPVSVIAGVQGALPPHRYSQAEVTDAFLAAPAFAEVGDLLRS
MMAR_2474|M.marinum_M ----------MSVIAGVFGALPPHRYSQSEITEAFLEFPGLQEHEEIVRR
MUL_1656|M.ulcerans_Agy99 ----------MSVIAGVFGALPPHRYSQSEITESFLEFPGLQEHEEIVRR
Mb1688|M.bovis_AF2122/97 ----------MSVIAGVFGALPPYRYSQRELTDSFVSIPDFEGYEDIVRQ
Rv1660|M.tuberculosis_H37Rv ----------MSVIAGVFGALPPYRYSQRELTDSFVSIPDFEGYEDIVRQ
MAV_3110|M.avium_104 ----------MSVIAGVFGALPPHRYPQRELTDFFVSIPEFEGYEDIVRQ
MSMEG_0808|M.smegmatis_MC2_155 MENTAFTPSGTCTIAGVHGVLPPHRYTQAEVTEALLDLPGYADFEGIVRS
MAB_1141c|M.abscessus_ATCC_199 ----------MTDIAEITGIN----------KIGLLSLGAYRP-ERVVTN
** : * . :: ::
Mvan_1196|M.vanbaalenii_PYR-1 LHKHAKVDARHFVLPMDRYPTLVDFGETNDLYLEHSLDLACEALTAALDE
Mflv_5140|M.gilvum_PYR-GCK LHTNAKVDARHLVLPLEQYPDLLDFGDANDLYIHHAVELGCQALTAALDE
MMAR_2474|M.marinum_M MHAAAKVNSRQFVLPIEQYPALTDFGDANEIFIEKAVDLGIEALMGALDE
MUL_1656|M.ulcerans_Agy99 MHAAAKVNSRQFVLPIEQYPALTDFGDTNEIFIEKAVDLGIEALMGALDE
Mb1688|M.bovis_AF2122/97 LHASAKVNSRHLVLPLEKYPKLTDFGEANKIFIEKAVDLGVQALAGALDE
Rv1660|M.tuberculosis_H37Rv LHASAKVNSRHLVLPLEKYPKLTDFGEANKIFIEKAVDLGVQALAGALDE
MAV_3110|M.avium_104 LHASAKVGSRHLVLPLEQYPTLTDFGVANRIFIDHAVTLGCAALSGALDE
MSMEG_0808|M.smegmatis_MC2_155 LHKSAKVNSRYLTLPLEEYAALDDFGETNGLFIEHATELGCAALTGALDE
MAB_1141c|M.abscessus_ATCC_199 DEICERIESSDEWIYTRTGIKTRRFAADEETAQTLAIEAGRKAIANALLT
. :: : : *. : : . *: **
Mvan_1196|M.vanbaalenii_PYR-1 AGLGPRDLDMIMTTTVTGIAVPSLDARIADRLGLRPDVRRVPLFGL--GC
Mflv_5140|M.gilvum_PYR-GCK AGLAPSDLDVIMSTTVTGIAVPSLDARIAAKLGLRPDVRRVPLFGL--GC
MMAR_2474|M.marinum_M AGLTPQDIDLIITTTVTGVAVPSLDARIAGRLGLRPDVRRMPLFGL--GC
MUL_1656|M.ulcerans_Agy99 AGLTPQDIDLIITTTVTGVAVPSLDARIAGRLGLRPDVRRMPLFGL--GC
Mb1688|M.bovis_AF2122/97 SGLRPEDLDVLITATVTGLAVPSLDARIAGRLGLRADVRRVPLFGL--GC
Rv1660|M.tuberculosis_H37Rv SGLRPEDLDVLITATVTGLAVPSLDARIAGRLGLRADVRRVPLFGL--GC
MAV_3110|M.avium_104 AGLKPEDLDVLITTTVTGLAVPSVDARIAARLGLRDDVRRVPLFGL--GC
MSMEG_0808|M.smegmatis_MC2_155 AGLRPEDVDLIITTTVTGAVVPSLDARIAGRVGLRPDVRRVPIFGL--GC
MAB_1141c|M.abscessus_ATCC_199 G---AEIDAVIVATSTHYLQTPASAPAVATALGAQG----VPAFDLSAGC
. . ::::::. .*: . :* :* : :* *.* **
Mvan_1196|M.vanbaalenii_PYR-1 VAGAAGIARMHDYLRGAPDHIAALVSVELCSLTYPAVEPTMASLVGSALF
Mflv_5140|M.gilvum_PYR-GCK VAGAAGIARMHDYLRGAPGHVAALISVELCSLTRPALAPTIGSLVGSALF
MMAR_2474|M.marinum_M VAGAAGVARLHDYLRGAPDGVAVLIAVELCSLTYPAVKPTVSGIVGTALF
MUL_1656|M.ulcerans_Agy99 VAGAAGVARLHDYLRGAPDGVAVLIAVELCSLTYPAVKPTVSGIVGTALF
Mb1688|M.bovis_AF2122/97 VAGAAGVARLHDYLRGAPDGVAALVSVELCSLTYPGYKPTLPGLVGSALF
Rv1660|M.tuberculosis_H37Rv VAGAAGVARLHDYLRGAPDGVAALVSVELCSLTYPGYKPTLPGLVGSALF
MAV_3110|M.avium_104 VAGAAGVARMHDYLRGAPDAVAALVSVELCSLTYPGYKPSLAGLVGSALF
MSMEG_0808|M.smegmatis_MC2_155 VAGAAGVARLNDYLRGAPDKVAVLVSVELCSLTH-KHTPSMPTLVAGALF
MAB_1141c|M.abscessus_ATCC_199 AGFGYAVGVAGDMIRGGSASKVLVIGSEKLSPALDMTDRGN-----CFIF
.. . .:. * :**.. . ::. * * : :*
Mvan_1196|M.vanbaalenii_PYR-1 GDGAAAVIAVGAERAETIGATGPEVLDTRSRMYPDSLDTMGWRVG-----
Mflv_5140|M.gilvum_PYR-GCK GDGAAAVVAVGEERAGHIRASGPDVLDSRSRMYPDSLDIMGWSVN-----
MMAR_2474|M.marinum_M GDGAAAVVAVGDRRAEQIRAGGPDILDSRSRLYPESLDIMGWDVG-----
MUL_1656|M.ulcerans_Agy99 GDGAAAVVAVGDRRAEQIRAGGPDILDSRSRLYPESLDIMGWDVG-----
Mb1688|M.bovis_AF2122/97 ADGAAAVVAAGVKRAQDIGADGPDILDSRSHLYPDSLRTMGYDVG-----
Rv1660|M.tuberculosis_H37Rv ADGAAAVVAAGVKRAQDIGADGPDILDSRSHLYPDSLRTMGYDVG-----
MAV_3110|M.avium_104 ADGAGAVVAVGERRAEQLDAAGPSVLDSRSHLYPDSLRTMGYDVG-----
MSMEG_0808|M.smegmatis_MC2_155 GDGASAVVAVGAGRAEQLDPIGPDVLDSRSHLYPDSLHAMGWDIG-----
MAB_1141c|M.abscessus_ATCC_199 GDGAG-AVVVGETAEQGIGPTVWGSDGSQSNAIRQDIDWMDFAAAPDPEG
.***. .:..* : . .::*. :.: *.:
Mvan_1196|M.vanbaalenii_PYR-1 ASGFQLILSPNLPELIERYLADDMAGFLGAHDLQIGDIAAWVSHPGGPKV
Mflv_5140|M.gilvum_PYR-GCK HNGFQLVLSPELPALIERYLPDDMASFLGEHDLGIGDIATWVSHPGGPKV
MMAR_2474|M.marinum_M SHGLQLRLAPELTNLIEEYLPDDVDAFLAMHDLTKNDIGAWVSHPGGPKV
MUL_1656|M.ulcerans_Agy99 SHGLQLRLAPELTNLIEEYLPDDVDAFLAMHDLTKNDIGAWVSHPGGPKV
Mb1688|M.bovis_AF2122/97 SAGFELVLSRDLAAVVEQYLGNDVTTFLASHGLSTTDVGAWVTHPGGPKI
Rv1660|M.tuberculosis_H37Rv SAGFELVLSRDLAAVVEQYLGNDVTTFLASHGLSTTDVGAWVTHPGGPKI
MAV_3110|M.avium_104 ATGFELVLSKDVAAVVEQYIEDDVTGFLGAHGLTTNDIGAFVSHPGGPKV
MSMEG_0808|M.smegmatis_MC2_155 TKGFEIVLSAEVPGFVGRYLGDDVTEFLGEHGVATDDVSAWVSHPGGPKV
MAB_1141c|M.abscessus_ATCC_199 PRPYLRIEGTSVFRWAAFEMGKVGQRAMEAAKVGPEQIDVFVPHQANTRI
. .: : . : : :: .:*.* ...::
Mvan_1196|M.vanbaalenii_PYR-1 MEAISETLGLPDDALDLAWRSLAEVGNLSSSSVLHVLRDTRAK-KPPAGT
Mflv_5140|M.gilvum_PYR-GCK LEAIASALDLPDDALELSWRSLAEIGNLSSSSVLHILRDTIAK-KPPAAT
MMAR_2474|M.marinum_M LDAITNSLGLPSDALELTWRSLGEIGNLSSASVLHILRDTIAK-VPPSGS
MUL_1656|M.ulcerans_Agy99 LDAIANSLGLPSDALELTWRSLGEIGNLSSASVLRILRDTIAK-VPPSGS
Mb1688|M.bovis_AF2122/97 INAITETLDLSPQALELTWRSLGEIGNLSSASVLHVLRDTIAK-PPPSGS
Rv1660|M.tuberculosis_H37Rv INAITETLDLSPQALELTWRSLGEIGNLSSASVLHVLRDTIAK-PPPSGS
MAV_3110|M.avium_104 IEAINAALGLPPEALELTWRSLGEIGNLSSASVLHVLRDTLAK-PPPSDS
MSMEG_0808|M.smegmatis_MC2_155 IEAIIETLDLPSDALDLTWQSLAEVGNLSSSSVLHVLRDTIRK-RPPAGT
MAB_1141c|M.abscessus_ATCC_199 NELLAKNLHLRADAVIAN--DIEHTGNTSAASVPLAMETLLSTGAAKPGD
: : * * :*: .: . ** *::** :. . . .
Mvan_1196|M.vanbaalenii_PYR-1 PGLLMAMGPGFCSELVLLRWR-----
Mflv_5140|M.gilvum_PYR-GCK PGLLMAMGPGFCSELVLLRWH-----
MMAR_2474|M.marinum_M PGLMLAMGPGFCAELVLLRWH-----
MUL_1656|M.ulcerans_Agy99 PGLMLAMGPGFCAELVLLRWH-----
Mb1688|M.bovis_AF2122/97 PGLMIAMGPGFCSELVLLRWH-----
Rv1660|M.tuberculosis_H37Rv PGLMIAMGPGFCSELVLLRWH-----
MAV_3110|M.avium_104 PGLMLAMGPGFCSELVLLRWH-----
MSMEG_0808|M.smegmatis_MC2_155 PGVLMAMGPGFCSELVLLRWR-----
MAB_1141c|M.abscessus_ATCC_199 LALLIGYGAGLSYAAQVVRMPPVPFE
.:::. *.*:. ::*