For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSVIAGVFGALPPYRYSQRELTDSFVSIPDFEGYEDIVRQLHASAKVNSRHLVLPLEKYPKLTDFGEANK IFIEKAVDLGVQALAGALDESGLRPEDLDVLITATVTGLAVPSLDARIAGRLGLRADVRRVPLFGLGCVA GAAGVARLHDYLRGAPDGVAALVSVELCSLTYPGYKPTLPGLVGSALFADGAAAVVAAGVKRAQDIGADG PDILDSRSHLYPDSLRTMGYDVGSAGFELVLSRDLAAVVEQYLGNDVTTFLASHGLSTTDVGAWVTHPGG PKIINAITETLDLSPQALELTWRSLGEIGNLSSASVLHVLRDTIAKPPPSGSPGLMIAMGPGFCSELVLL RWH
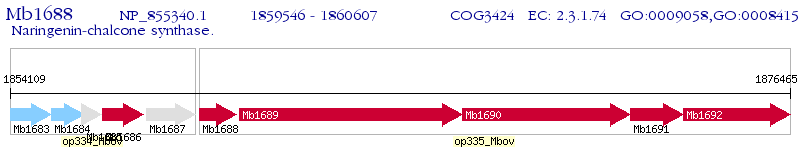
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1688 | pks10 | - | 100% (353) | chalcone synthase |
| M. bovis AF2122 / 97 | Mb1693 | pks11 | e-153 | 74.15% (352) | chalcone synthase |
| M. bovis AF2122 / 97 | Mb1406 | - | 7e-30 | 26.89% (357) | hypothetical protein Mb1406 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_5140 | - | 1e-138 | 67.42% (353) | chalcone/stilbene synthase domain-containing protein |
| M. tuberculosis H37Rv | Rv1660 | pks10 | 0.0 | 100.00% (353) | chalcone synthase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_2470 | pks10 | 1e-178 | 86.12% (353) | chalcone synthase, Pks10 |
| M. avium 104 | MAV_3110 | - | 1e-168 | 82.15% (353) | Pks10 protein |
| M. smegmatis MC2 155 | MSMEG_0808 | - | 1e-138 | 67.91% (349) | chalcone synthase Pks10 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_1656 | pks11 | 1e-151 | 72.24% (353) | chalcone synthase, Pks11 |
| M. vanbaalenii PYR-1 | Mvan_1196 | - | 1e-141 | 66.76% (352) | chalcone/stilbene synthase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_5140|M.gilvum_PYR-GCK ---MTDVHTPVSVIAGVQGALPPHRYSQAEVTDAFLAAPAFAEVGDLLRS
Mvan_1196|M.vanbaalenii_PYR-1 ------MNDRISVIAGVQGALPPYRYSQAEITERFVAVPAFAELEDVVRG
Mb1688|M.bovis_AF2122/97 ----------MSVIAGVFGALPPYRYSQRELTDSFVSIPDFEGYEDIVRQ
Rv1660|M.tuberculosis_H37Rv ----------MSVIAGVFGALPPYRYSQRELTDSFVSIPDFEGYEDIVRQ
MMAR_2470|M.marinum_M ----------MSVIAGVFGALPPHRYSQRELTETFVNIPDFEGYEELVRQ
MAV_3110|M.avium_104 ----------MSVIAGVFGALPPHRYPQRELTDFFVSIPEFEGYEDIVRQ
MUL_1656|M.ulcerans_Agy99 ----------MSVIAGVFGALPPHRYSQSEITESFLEFPGLQEHEEIVRR
MSMEG_0808|M.smegmatis_MC2_155 MENTAFTPSGTCTIAGVHGVLPPHRYTQAEVTEALLDLPGYADFEGIVRS
..**** *.***:**.* *:*: :: * ::*
Mflv_5140|M.gilvum_PYR-GCK LHTNAKVDARHLVLPLEQYPDLLDFGDANDLYIHHAVELGCQALTAALDE
Mvan_1196|M.vanbaalenii_PYR-1 LHKHAKVDARHFVLPMDRYPTLVDFGETNDLYLEHSLDLACEALTAALDE
Mb1688|M.bovis_AF2122/97 LHASAKVNSRHLVLPLEKYPKLTDFGEANKIFIEKAVDLGVQALAGALDE
Rv1660|M.tuberculosis_H37Rv LHASAKVNSRHLVLPLEKYPKLTDFGEANKIFIEKAVDLGVQALAGALDE
MMAR_2470|M.marinum_M LHANAKVNSRHLVLPLETYPKLNDFGEANGIFIDKAVDLGVEALMGALDE
MAV_3110|M.avium_104 LHASAKVGSRHLVLPLEQYPTLTDFGVANRIFIDHAVTLGCAALSGALDE
MUL_1656|M.ulcerans_Agy99 MHAAAKVNSRQFVLPIEQYPALTDFGDTNEIFIEKAVDLGIEALMGALDE
MSMEG_0808|M.smegmatis_MC2_155 LHKSAKVNSRYLTLPLEEYAALDDFGETNGLFIEHATELGCAALTGALDE
:* ***.:* :.**:: *. * *** :* :::.:: *. ** .****
Mflv_5140|M.gilvum_PYR-GCK AGLAPSDLDVIMSTTVTGIAVPSLDARIAAKLGLRPDVRRVPLFGLGCVA
Mvan_1196|M.vanbaalenii_PYR-1 AGLGPRDLDMIMTTTVTGIAVPSLDARIADRLGLRPDVRRVPLFGLGCVA
Mb1688|M.bovis_AF2122/97 SGLRPEDLDVLITATVTGLAVPSLDARIAGRLGLRADVRRVPLFGLGCVA
Rv1660|M.tuberculosis_H37Rv SGLRPEDLDVLITATVTGLAVPSLDARIAGRLGLRADVRRVPLFGLGCVA
MMAR_2470|M.marinum_M AGLQPEDLDVLITTTVTGLAVPSLDARIAGRVGLRPDVRRVPLFGLGCVA
MAV_3110|M.avium_104 AGLKPEDLDVLITTTVTGLAVPSVDARIAARLGLRDDVRRVPLFGLGCVA
MUL_1656|M.ulcerans_Agy99 AGLTPQDIDLIITTTVTGVAVPSLDARIAGRLGLRPDVRRMPLFGLGCVA
MSMEG_0808|M.smegmatis_MC2_155 AGLRPEDVDLIITTTVTGAVVPSLDARIAGRVGLRPDVRRVPIFGLGCVA
:** * *:*:::::**** .***:***** ::*** ****:*:*******
Mflv_5140|M.gilvum_PYR-GCK GAAGIARMHDYLRGAPGHVAALISVELCSLTRPALAPTIGSLVGSALFGD
Mvan_1196|M.vanbaalenii_PYR-1 GAAGIARMHDYLRGAPDHIAALVSVELCSLTYPAVEPTMASLVGSALFGD
Mb1688|M.bovis_AF2122/97 GAAGVARLHDYLRGAPDGVAALVSVELCSLTYPGYKPTLPGLVGSALFAD
Rv1660|M.tuberculosis_H37Rv GAAGVARLHDYLRGAPDGVAALVSVELCSLTYPGYKPTLPGLVGSALFAD
MMAR_2470|M.marinum_M GAAGVARLHDYLRGAPNGIAALISVELCSLTFPGYKPSAAGLVGSALFAD
MAV_3110|M.avium_104 GAAGVARMHDYLRGAPDAVAALVSVELCSLTYPGYKPSLAGLVGSALFAD
MUL_1656|M.ulcerans_Agy99 GAAGVARLHDYLRGAPDGVAVLIAVELCSLTYPAVKPTVSGIVGTALFGD
MSMEG_0808|M.smegmatis_MC2_155 GAAGVARLNDYLRGAPDKVAVLVSVELCSLTH-KHTPSMPTLVAGALFGD
****:**::*******. :*.*::******* *: :*. ***.*
Mflv_5140|M.gilvum_PYR-GCK GAAAVVAVGEERAGHIRASGPDVLDSRSRMYPDSLDIMGWSVNHNGFQLV
Mvan_1196|M.vanbaalenii_PYR-1 GAAAVIAVGAERAETIGATGPEVLDTRSRMYPDSLDTMGWRVGASGFQLI
Mb1688|M.bovis_AF2122/97 GAAAVVAAGVKRAQDIGADGPDILDSRSHLYPDSLRTMGYDVGSAGFELV
Rv1660|M.tuberculosis_H37Rv GAAAVVAAGVKRAQDIGADGPDILDSRSHLYPDSLRTMGYDVGSAGFELV
MMAR_2470|M.marinum_M GAAAVVAVGDGRAEEVGASGPDILDSCSHLYPDSLRTMGYDVGSAGFELV
MAV_3110|M.avium_104 GAGAVVAVGERRAEQLDAAGPSVLDSRSHLYPDSLRTMGYDVGATGFELV
MUL_1656|M.ulcerans_Agy99 GAAAVVAVGDRRAEQIRAGGPDILDSRSRLYPESLDIMGWDVGSHGLQLR
MSMEG_0808|M.smegmatis_MC2_155 GASAVVAVGAGRAEQLDPIGPDVLDSRSHLYPDSLHAMGWDIGTKGFEIV
**.**:*.* ** : . **.:**: *::**:** **: :. *:::
Mflv_5140|M.gilvum_PYR-GCK LSPELPALIERYLPDDMASFLGEHDLGIGDIATWVSHPGGPKVLEAIASA
Mvan_1196|M.vanbaalenii_PYR-1 LSPNLPELIERYLADDMAGFLGAHDLQIGDIAAWVSHPGGPKVMEAISET
Mb1688|M.bovis_AF2122/97 LSRDLAAVVEQYLGNDVTTFLASHGLSTTDVGAWVTHPGGPKIINAITET
Rv1660|M.tuberculosis_H37Rv LSRDLAAVVEQYLGNDVTTFLASHGLSTTDVGAWVTHPGGPKIINAITET
MMAR_2470|M.marinum_M LSKDIADVVEEYVAEDVTTFLAGHGLSVTDIGAWVTHPGGPKIINAITES
MAV_3110|M.avium_104 LSKDVAAVVEQYIEDDVTGFLGAHGLTTNDIGAFVSHPGGPKVIEAINAA
MUL_1656|M.ulcerans_Agy99 LAPELTNLIEEYLPDDVDAFLAMHDLTKNDIGAWVSHPGGPKVLDAIANS
MSMEG_0808|M.smegmatis_MC2_155 LSAEVPGFVGRYLGDDVTEFLGEHGVATDDVSAWVSHPGGPKVIEAIIET
*: ::. .: .*: :*: **. *.: *:.::*:******:::** :
Mflv_5140|M.gilvum_PYR-GCK LDLPDDALELSWRSLAEIGNLSSSSVLHILRDTIAKKPPAATPGLLMAMG
Mvan_1196|M.vanbaalenii_PYR-1 LGLPDDALDLAWRSLAEVGNLSSSSVLHVLRDTRAKKPPAGTPGLLMAMG
Mb1688|M.bovis_AF2122/97 LDLSPQALELTWRSLGEIGNLSSASVLHVLRDTIAKPPPSGSPGLMIAMG
Rv1660|M.tuberculosis_H37Rv LDLSPQALELTWRSLGEIGNLSSASVLHVLRDTIAKPPPSGSPGLMIAMG
MMAR_2470|M.marinum_M LNLPPEALELTWRSLGEIGNLSSASVLHVLRDTIAKPPPSGSPGLMMAMG
MAV_3110|M.avium_104 LGLPPEALELTWRSLGEIGNLSSASVLHVLRDTLAKPPPSDSPGLMLAMG
MUL_1656|M.ulcerans_Agy99 LGLPSDALELTWRSLGEIGNLSSASVLRILRDTIAKVPPSGSPGLMLAMG
MSMEG_0808|M.smegmatis_MC2_155 LDLPSDALDLTWQSLAEVGNLSSSSVLHVLRDTIRKRPPAGTPGVLMAMG
*.*. :**:*:*:**.*:*****:***::**** * **: :**:::***
Mflv_5140|M.gilvum_PYR-GCK PGFCSELVLLRWH
Mvan_1196|M.vanbaalenii_PYR-1 PGFCSELVLLRWR
Mb1688|M.bovis_AF2122/97 PGFCSELVLLRWH
Rv1660|M.tuberculosis_H37Rv PGFCSELVLLRWH
MMAR_2470|M.marinum_M PGFCSELVLLRWH
MAV_3110|M.avium_104 PGFCSELVLLRWH
MUL_1656|M.ulcerans_Agy99 PGFCAELVLLRWH
MSMEG_0808|M.smegmatis_MC2_155 PGFCSELVLLRWR
****:*******: