For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PLEDAMRTQRSIRRLKSDPVDDSLVLHLLELAMKAPTGSNAQNWEFIVVKDREVVAKLGRLNRRAMQLFG PIYRRSIARRGDEKMLRLQKAVQWQADHFEEIPVVVVACLRGVIAPWPRISTSSAYGSIYPAVQNLLLAA RAAGLGAGLITVPLWSTVLARRALGLPWNVTPCAVVPLGWPIGKYGPTTRRPVAEIVSLDRYGNRAFR*
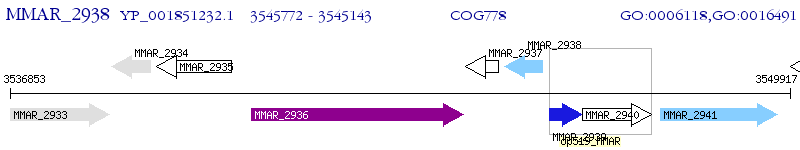
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_2938 | - | - | 100% (209) | nitroreductase |
| M. marinum M | MMAR_4721 | - | 2e-29 | 37.20% (207) | nitroreductase |
| M. marinum M | MMAR_1160 | - | 1e-16 | 29.06% (203) | nitroreductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3403c | - | 2e-17 | 29.61% (206) | oxidoreductase |
| M. gilvum PYR-GCK | Mflv_3382 | - | 1e-65 | 57.28% (213) | nitroreductase |
| M. tuberculosis H37Rv | Rv3368c | - | 2e-17 | 29.61% (206) | oxidoreductase |
| M. leprae Br4923 | MLBr_00418 | - | 1e-17 | 29.38% (211) | putative oxidoreductase |
| M. abscessus ATCC 19977 | MAB_3693c | - | 9e-22 | 31.10% (209) | oxidoreductase |
| M. avium 104 | MAV_0933 | - | 1e-28 | 38.12% (202) | nitroreductase family protein |
| M. smegmatis MC2 155 | MSMEG_6627 | - | 2e-20 | 34.18% (196) | nitroreductase family protein |
| M. thermoresistible (build 8) | TH_2448 | - | 1e-71 | 62.50% (216) | PUTATIVE nitroreductase |
| M. ulcerans Agy99 | MUL_2198 | - | 1e-117 | 99.52% (209) | nitroreductase |
| M. vanbaalenii PYR-1 | Mvan_3115 | - | 2e-67 | 59.53% (215) | nitroreductase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2938|M.marinum_M -------------------MPLEDAMRTQRSIRR--LKSDPVDDSLVLHL
MUL_2198|M.ulcerans_Agy99 -------------------MPLEDAMRTQRSIRR--LKSDPVDDSLVLHL
TH_2448|M.thermoresistible__bu MPDTPKPIPSVAEALARLDMPLAEAMMTQRAVRR--VLPDPVDDELVLKC
Mvan_3115|M.vanbaalenii_PYR-1 ---MPEPIPSAAEALARLDMPLVEAMMTQRAIRR--VLPDPVDDEIVLKC
Mflv_3382|M.gilvum_PYR-GCK ---MADPTPSTSEALARLDMSLIDVMMTQRAVRR--VYPDPVDDEIVMKC
MAV_0933|M.avium_104 ------------------------MMSTARTIRR--FTDEPVDDATLARC
MSMEG_6627|M.smegmatis_MC2_155 --------------MPQPTNDVWEVLSTARSIRR--FTDEPVDDATLHRC
Mb3403c|M.bovis_AF2122/97 ---------------MTLNLSVDEVLTTTRSVRKRLDFDKPVPRDVLMEC
Rv3368c|M.tuberculosis_H37Rv ---------------MTLNLSVDEVLTTTRSVRKRLDFDKPVPRDVLMEC
MLBr_00418|M.leprae_Br4923 -------------------MSVDEVLTTTRSVRKRLDFDKPVPRDVLMEC
MAB_3693c|M.abscessus_ATCC_199 --------------MTSLNLSADEVLTTTRSVRKRLDFDKPVERAVVEEC
: * *::*: .** : .
MMAR_2938|M.marinum_M LELAMKAPTGSNAQNWEFIVVKDREVVAKLGRLNRRAMQLFGPIYRRSIA
MUL_2198|M.ulcerans_Agy99 LELAMKAPTGSNAQNWEFIVVKDREVVAKLGRLNRRAMQLFGPIYRRSIA
TH_2448|M.thermoresistible__bu IELALRAPTGSNGQNWEFIVVKDPAVKAKLADRYRQAWAVYGGIGRR-VT
Mvan_3115|M.vanbaalenii_PYR-1 LELALRAPTGANGQNWEFVVVKDPKIKRKLQRRYRLAWRVFHRTSIRDIA
Mflv_3382|M.gilvum_PYR-GCK LEVALHAPTGANGQNWEFIVVKDYETKKKLAKRYRQAWNLYYQTVIRRVS
MAV_0933|M.avium_104 LGAARWAPSGANAQGWRFVVLRSPEQRAVVAKAAAQALEVIEPVYGMSRP
MSMEG_6627|M.smegmatis_MC2_155 LQAAAWAPNGANAQLWRFVVLDAPEQRAAVAGAARSALATIESIYGMSRP
Mb3403c|M.bovis_AF2122/97 LELALQAPTGSNSQGWQWVFVEDAAKKKAIADVYLANARGYLSGPAPEYP
Rv3368c|M.tuberculosis_H37Rv LELALQAPTGSNSQGWQWVFVEDAAKKKAIADVYLANARGYLSGPAPEYP
MLBr_00418|M.leprae_Br4923 LQLALQAPTGSNSQGWHWVFVEDAEKRKAIGDIYLVNARCYLSQPAPEYP
MAB_3693c|M.abscessus_ATCC_199 LNIAMQAPTGSNHQGWHWVIVEDAEKKKAIADIYREGWNKYAKGAGATYA
: * **.*:* * *.::.: : .
MMAR_2938|M.marinum_M RRG--DEKMLRLQKAVQWQADHFEEIPVVVVACLRG-------VIAPWPR
MUL_2198|M.ulcerans_Agy99 RRG--DEKMLRLQKAVQWQADHFEEIPVVVVACLRG-------VIAPWPR
TH_2448|M.thermoresistible__bu AG---DESMAKILRAVQWQVDHFTEIPVLVVACLRAGPREGRLPFMPAPP
Mvan_3115|M.vanbaalenii_PYR-1 SY---DEEMAKSVRAIEWQLEHFHEIPVLVIACLRLSAREGKVPYVPMPH
Mflv_3382|M.gilvum_PYR-GCK KR---DPSMAKTARAVQWQVDHFTEVPVLIVACLKLGPKDGRIPFVPMPH
MAV_0933|M.avium_104 APDD-HSRRARTYRATYELHDRAGEFTSVLFAQAHY------------PT
MSMEG_6627|M.smegmatis_MC2_155 ADDD-HSRAARNNRATYELHDRAGELTSVLFVAYKN------------EF
Mb3403c|M.bovis_AF2122/97 DGDTRGERMGRVRDSATYLAEHMHRAPVLLIPCLKGRED--------ESA
Rv3368c|M.tuberculosis_H37Rv DGDTRGERMGRVRDSATYLAEHMHRAPVLLIPCLKGRED--------ESA
MLBr_00418|M.leprae_Br4923 EGDTRGERMRLVRDSATYLAEHMHEVPVLLIPCLLGRAE--------ESP
MAB_3693c|M.abscessus_ATCC_199 EGDTRAERKAKVVDSAGYLAENFERSPLFLIPCIEGRLDN-------LPV
: :. . . .:.
MMAR_2938|M.marinum_M ISTSSAYGSIYPAVQNLLLAARAAGLGAGLITVPLWS--TVLARRALGLP
MUL_2198|M.ulcerans_Agy99 ISTSSAYGSIYPAVQNLLLAARAAGLGAGLITVPLWS--TVLARRALGLP
TH_2448|M.thermoresistible__bu VAASSFFGSIYPSVQNLLLAARAMGLGASLITLPLWS--MTSARRILKLP
Mvan_3115|M.vanbaalenii_PYR-1 AAASAFFGSIYPSVQNLLLAARAMGLGASLITLPLWN--LTSARRVLKLP
Mflv_3382|M.gilvum_PYR-GCK AALSGFYGSIYPSVQNLLLAARAMGLGASLITLPLWS--VTSTRKTLGLP
MAV_0933|M.avium_104 ASELLLGGSIFPAMQNFLLAARAQGLGACLTSWASYGG-EQLLREAVGVP
MSMEG_6627|M.smegmatis_MC2_155 ASDYLQAGSIYPAMQNFYLAARAQGLGACFTSWAAYQG-EQILRDAVGVP
Mb3403c|M.bovis_AF2122/97 VGGVSFWASLFPAVWSFCLALRSRGLGSCWTTLHLLDNGEHKVADVLGIP
Rv3368c|M.tuberculosis_H37Rv VGGVSFWASLFPAVWSFCLALRSRGLGSCWTTLHLLDNGEHKVADVLGIP
MLBr_00418|M.leprae_Br4923 LGAVSYWASLFPAVWSFCLALRSRGLGTCWTSLHLLGDGEQRAAEVLGIP
MAB_3693c|M.abscessus_ATCC_199 VGAASSWGSLLPAVWSFMLAARNRGLGSAWTTLHLMDDGEKRTADIVGIP
. .*: *:: .: ** * ***: : : :*
MMAR_2938|M.marinum_M -WNVTPCAVVPLGWPIGK-YGPTTRRPVAEIVSLDRYGNRAFR-------
MUL_2198|M.ulcerans_Agy99 -RNVTPCAVVPLGWPIGK-YGPTTRRPVAEIVSLDRYGNRAFR-------
TH_2448|M.thermoresistible__bu -LSVTPCCIVPLGWPRGR-YGPTTRRPVREVVHLDSYGNRAWQSPS----
Mvan_3115|M.vanbaalenii_PYR-1 -TEVTPCCVVPLGWPRGR-YGPTTRRPVDEVVHLDRYGNRAWFGPRGADR
Mflv_3382|M.gilvum_PYR-GCK -VSVTPCCVVPLGWPRGR-YGPTTRRPVAEVMHLNTYGNRPWMGTG----
MAV_0933|M.avium_104 -EGWMIAGHIVVGWPKGK-HGPLRRRPLAEFVNLDRWDAPADGLLSGL--
MSMEG_6627|M.smegmatis_MC2_155 -AEWFLAGHVVIGRPRGR-HGPVRRRPLDDVVFRNTWDAARADIVYGQGA
Mb3403c|M.bovis_AF2122/97 YDEYSQGGLLPIAYTQGIDFRPAKRLPAESVTHWNGW-------------
Rv3368c|M.tuberculosis_H37Rv YDEYSQGGLLPIAYTQGIDFRPAKRLPAESVTHWNGW-------------
MLBr_00418|M.leprae_Br4923 SDKYSQGGLFPIAYTKGTDFRPANRLPAENVTHWDIW-------------
MAB_3693c|M.abscessus_ATCC_199 FDKVTQGGLFPIAYTVGTDFRLAKRQPLADVLHWDTW-------------
. :. . * . * * .. : :
MMAR_2938|M.marinum_M -----
MUL_2198|M.ulcerans_Agy99 -----
TH_2448|M.thermoresistible__bu -----
Mvan_3115|M.vanbaalenii_PYR-1 P----
Mflv_3382|M.gilvum_PYR-GCK -----
MAV_0933|M.avium_104 -----
MSMEG_6627|M.smegmatis_MC2_155 RPSRT
Mb3403c|M.bovis_AF2122/97 -----
Rv3368c|M.tuberculosis_H37Rv -----
MLBr_00418|M.leprae_Br4923 -----
MAB_3693c|M.abscessus_ATCC_199 -----