For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPQPTNDVWEVLSTARSIRRFTDEPVDDATLHRCLQAAAWAPNGANAQLWRFVVLDAPEQRAAVAGAARS ALATIESIYGMSRPADDDHSRAARNNRATYELHDRAGELTSVLFVAYKNEFASDYLQAGSIYPAMQNFYL AARAQGLGACFTSWAAYQGEQILRDAVGVPAEWFLAGHVVIGRPRGRHGPVRRRPLDDVVFRNTWDAARA DIVYGQGARPSRT
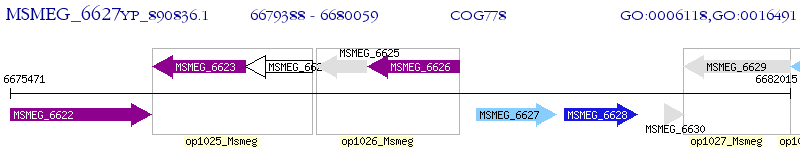
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6627 | - | - | 100% (223) | nitroreductase family protein |
| M. smegmatis MC2 155 | MSMEG_1511 | - | 7e-15 | 33.80% (213) | putative oxidoreductase |
| M. smegmatis MC2 155 | MSMEG_5155 | - | 1e-12 | 27.36% (212) | nitroreductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3403c | - | 2e-14 | 31.60% (212) | oxidoreductase |
| M. gilvum PYR-GCK | Mflv_1262 | - | 1e-106 | 81.82% (220) | nitroreductase |
| M. tuberculosis H37Rv | Rv3368c | - | 2e-14 | 31.60% (212) | oxidoreductase |
| M. leprae Br4923 | MLBr_00418 | - | 9e-16 | 30.19% (212) | putative oxidoreductase |
| M. abscessus ATCC 19977 | MAB_3693c | - | 3e-15 | 32.39% (213) | oxidoreductase |
| M. marinum M | MMAR_4721 | - | 2e-77 | 70.77% (195) | nitroreductase |
| M. avium 104 | MAV_0933 | - | 3e-78 | 68.78% (205) | nitroreductase family protein |
| M. thermoresistible (build 8) | TH_1618 | - | 1e-111 | 85.14% (222) | nitroreductase family protein |
| M. ulcerans Agy99 | MUL_2198 | - | 8e-21 | 34.18% (196) | nitroreductase |
| M. vanbaalenii PYR-1 | Mvan_5547 | - | 1e-101 | 78.28% (221) | nitroreductase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1262|M.gilvum_PYR-GCK --------------MDMPHPTDNVWEVLSTARSIRR--FTDEPVDSATLD
Mvan_5547|M.vanbaalenii_PYR-1 MAKMLDICLRVLYTIDMPHPTDDVWEVLSTARSIRR--FTDEPVTDAVID
TH_1618|M.thermoresistible__bu ----------------MPHPTNDVWEVLSTARSIRR--FTDEPVDDATLQ
MSMEG_6627|M.smegmatis_MC2_155 ----------------MPQPTNDVWEVLSTARSIRR--FTDEPVDDATLH
MMAR_4721|M.marinum_M ---------------------------MSTARTVRR--FTDRPVDDATLT
MAV_0933|M.avium_104 --------------------------MMSTARTIRR--FTDEPVDDATLA
MUL_2198|M.ulcerans_Agy99 ---------------------MPLEDAMRTQRSIRR--LKSDPVDDSLVL
Mb3403c|M.bovis_AF2122/97 -----------------MTLNLSVDEVLTTTRSVRKRLDFDKPVPRDVLM
Rv3368c|M.tuberculosis_H37Rv -----------------MTLNLSVDEVLTTTRSVRKRLDFDKPVPRDVLM
MLBr_00418|M.leprae_Br4923 ---------------------MSVDEVLTTTRSVRKRLDFDKPVPRDVLM
MAB_3693c|M.abscessus_ATCC_199 ----------------MTSLNLSADEVLTTTRSVRKRLDFDKPVERAVVE
: * *::*: . ** :
Mflv_1262|M.gilvum_PYR-GCK RCLQAATWAPNGANAQLWRFIVLDGPEQRAAVAQAAQIALQSIESIYGMT
Mvan_5547|M.vanbaalenii_PYR-1 RCLEAATWAPNGANAQLWRFIVLDGPEQRAAVGKAAQIALTSIEAIYGMT
TH_1618|M.thermoresistible__bu RCLQAATWAPNGANAQLWRFIVLDAPEQRAAVAEAAQIALHSIESIYGMS
MSMEG_6627|M.smegmatis_MC2_155 RCLQAAAWAPNGANAQLWRFVVLDAPEQRAAVAGAARSALATIESIYGMS
MMAR_4721|M.marinum_M RCLEAATWAPSGANAQGWRFVVLRSPEQRAVVAAAAARALAVIEPVYGMR
MAV_0933|M.avium_104 RCLGAARWAPSGANAQGWRFVVLRSPEQRAVVAKAAAQALEVIEPVYGMS
MUL_2198|M.ulcerans_Agy99 HLLELAMKAPTGSNAQNWEFIVVKDREVVAKLGRLNRRAMQLFGPIYRRS
Mb3403c|M.bovis_AF2122/97 ECLELALQAPTGSNSQGWQWVFVEDAAKKKAIADVYLANARGYLSGPAPE
Rv3368c|M.tuberculosis_H37Rv ECLELALQAPTGSNSQGWQWVFVEDAAKKKAIADVYLANARGYLSGPAPE
MLBr_00418|M.leprae_Br4923 ECLQLALQAPTGSNSQGWHWVFVEDAEKRKAIGDIYLVNARCYLSQPAPE
MAB_3693c|M.abscessus_ATCC_199 ECLNIAMQAPTGSNHQGWHWVIVEDAEKKKAIADIYREGWNKYAKGAGAT
. * * **.*:* * *.::.: :.
Mflv_1262|M.gilvum_PYR-GCK RP-APEDDSRSARNNRATYELHDRAGEFTSVLFVAYKNEFS-----SEFL
Mvan_5547|M.vanbaalenii_PYR-1 RP-DDDDASRAARNNRATYELHDRAEEYTSVLFTAFKNEFS-----SEFL
TH_1618|M.thermoresistible__bu RP-ADDDRSRAARNNRATYELHDRAGEYTSVLFTAFKNEFS-----SELL
MSMEG_6627|M.smegmatis_MC2_155 RP-ADDDHSRAARNNRATYELHDRAGELTSVLFVAYKNEFA-----SDYL
MMAR_4721|M.marinum_M RP-AADDDSRRARTYRATYELHDRAGEFTSVLFAQQHYPTA-----SELL
MAV_0933|M.avium_104 RP-APDDHSRRARTYRATYELHDRAGEFTSVLFAQAHYPTA-----SELL
MUL_2198|M.ulcerans_Agy99 IA-RRGDE-KMLRLQKAVQWQADHFEEIPVVVVACLRGVIAPWPRISTSS
Mb3403c|M.bovis_AF2122/97 YPDGDTRGERMGRVRDSATYLAEHMHRAPVLLIPCLKGRED-ESAVGGVS
Rv3368c|M.tuberculosis_H37Rv YPDGDTRGERMGRVRDSATYLAEHMHRAPVLLIPCLKGRED-ESAVGGVS
MLBr_00418|M.leprae_Br4923 YPEGDTRGERMRLVRDSATYLAEHMHEVPVLLIPCLLGRAE-ESPLGAVS
MAB_3693c|M.abscessus_ATCC_199 YAEGDTRAERKAKVVDSAGYLAENFERSPLFLIPCIEGRLDNLPVVGAAS
. : :. :. . . .:. .
Mflv_1262|M.gilvum_PYR-GCK QGGSIYPALQNFYLAARAQGLGACITSWASYGGE-QALREAVGIP-DEWF
Mvan_5547|M.vanbaalenii_PYR-1 QGGSIYPAIQNFYLAARAQGLGACLTSWASYAGGGQVLREAVGIP-DEWF
TH_1618|M.thermoresistible__bu QGGSIYPAMQNFYLAARAQGLGACFTSWASYEGE-KVLREAVGVP-DDWF
MSMEG_6627|M.smegmatis_MC2_155 QAGSIYPAMQNFYLAARAQGLGACFTSWAAYQGE-QILRDAVGVP-AEWF
MMAR_4721|M.marinum_M LGGSIFPAMQNFLLAARAQGLGACMTSWASYGGE-RLLREAVGVP-EDWL
MAV_0933|M.avium_104 LGGSIFPAMQNFLLAARAQGLGACLTSWASYGGE-QLLREAVGVP-EGWM
MUL_2198|M.ulcerans_Agy99 AYGSIYPAVQNLLLAARAAGLGAGLITVPLWSTV--LARRALGLP-RNVT
Mb3403c|M.bovis_AF2122/97 FWASLFPAVWSFCLALRSRGLGSCWTTLHLLDNGEHKVADVLGIPYDEYS
Rv3368c|M.tuberculosis_H37Rv FWASLFPAVWSFCLALRSRGLGSCWTTLHLLDNGEHKVADVLGIPYDEYS
MLBr_00418|M.leprae_Br4923 YWASLFPAVWSFCLALRSRGLGTCWTSLHLLGDGEQRAAEVLGIPSDKYS
MAB_3693c|M.abscessus_ATCC_199 SWGSLLPAVWSFMLAARNRGLGSAWTTLHLMDDGEKRTADIVGIPFDKVT
.*: **: .: ** * ***: : :*:*
Mflv_1262|M.gilvum_PYR-GCK LAGHVVVGWPRGK-HGPVRRRPLSDVVFRNHWDPERADITYGRGARPRGK
Mvan_5547|M.vanbaalenii_PYR-1 LAGHIVVGWPRGH-HGPVRRRPLTDVVFRNRWDPERADITYGRGARPRGK
TH_1618|M.thermoresistible__bu LAGHVVVGWPRGR-HGPVRRRPLGDVVFRNRWDPDRADITYGRGARPKRR
MSMEG_6627|M.smegmatis_MC2_155 LAGHVVIGRPRGR-HGPVRRRPLDDVVFRNTWDAARADIVYGQGARPSRT
MMAR_4721|M.marinum_M LAGHIVIGWPQGK-HGPLRRRPLSDVVNLDRWDRPAFPGLGS--------
MAV_0933|M.avium_104 IAGHIVVGWPKGK-HGPLRRRPLAEFVNLDRWDAPADGLLSGL-------
MUL_2198|M.ulcerans_Agy99 PCAVVPLGWPIGK-YGPTTRRPVAEIVSLDRYGNRAFR------------
Mb3403c|M.bovis_AF2122/97 QGGLLPIAYTQGIDFRPAKRLPAESVTHWNGW------------------
Rv3368c|M.tuberculosis_H37Rv QGGLLPIAYTQGIDFRPAKRLPAESVTHWNGW------------------
MLBr_00418|M.leprae_Br4923 QGGLFPIAYTKGTDFRPANRLPAENVTHWDIW------------------
MAB_3693c|M.abscessus_ATCC_199 QGGLFPIAYTVGTDFRLAKRQPLADVLHWDTW------------------
. . :. . * . * * .. : :
Mflv_1262|M.gilvum_PYR-GCK -
Mvan_5547|M.vanbaalenii_PYR-1 S
TH_1618|M.thermoresistible__bu -
MSMEG_6627|M.smegmatis_MC2_155 -
MMAR_4721|M.marinum_M -
MAV_0933|M.avium_104 -
MUL_2198|M.ulcerans_Agy99 -
Mb3403c|M.bovis_AF2122/97 -
Rv3368c|M.tuberculosis_H37Rv -
MLBr_00418|M.leprae_Br4923 -
MAB_3693c|M.abscessus_ATCC_199 -