For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TTAAMTEATSEAATYDVAIIGYGPTGATAANLLGQLGMKVVVIERDPDIYGRARAISTDEEVMRIWQSVG LAERLQSDMLPDRPIAFVDAHGVPFIEMKIEPRGSGHPPQQFLYQPAVDQVLRAGVARFANVEVLVEHEC LRVLNNDAAAELLLADLRTDTFKRLRARYVIAADGGSSPTRGQLGVGYTGRTYAERWVVIDTKVRRAWDA HDRLRFHCDPARPTVDCPTPLGHHRWEYPASADEADQKLVREEAIWRVLNAQGITDNEVEILRAVIYCHH VRVADRWRIGRVFLAGDAAHAMPPWIGQGMSAGVRDAANLCWKLAAVLNGNAPDSLLDSYQDERKPHVTE VTRRAVFTGRLITERRRAVAAVRNGLGRALTRFPSAMARLQRAMWIPPARYQDGFLARGHRAVGWQIPQP WVIDGNGATARLDDILGSQWVLLHTGDAPTGSQAWTQRGVPAVRLGPPESGARPDGIRDSGGTLLRWLAR KHASAVVLRPDGFVYAAAESGSPLPPPPAGHFGNTLPTSMKIEALA*
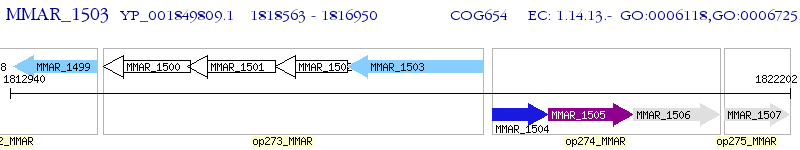
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_1503 | - | - | 100% (537) | FAD-dependent oxidoreductase |
| M. marinum M | MMAR_4777 | mhpA | 1e-68 | 33.14% (513) | hydroxylase |
| M. marinum M | MMAR_2987 | mhpA | 3e-59 | 29.62% (503) | 3-(3-hydroxy-phenyl) propionate hydroxylase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2401 | mhpA | 4e-67 | 33.14% (516) | 3-(3-hydroxyphenyl)propionate hydroxylase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2573c | mhpA | 6e-67 | 32.56% (516) | 3-(3-hydroxyphenyl)propionate hydroxylase |
| M. avium 104 | MAV_2518 | - | 1e-149 | 69.29% (368) | monooxygenase, FAD-binding |
| M. smegmatis MC2 155 | MSMEG_4866 | mhpA | 3e-65 | 32.26% (530) | 3-(3-hydroxyphenyl)propionate hydroxylase |
| M. thermoresistible (build 8) | TH_0968 | - | 1e-68 | 33.92% (510) | 3-(3-hydroxy-phenyl)propionate hydroxylase |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_0593 | - | 0.0 | 62.23% (511) | monooxygenase, FAD-binding |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1503|M.marinum_M MTTAAMTEATSEAATYDVAIIGYGPTGATAANLLGQLGMKVVVIERDPDI
MAV_2518|M.avium_104 --------------------------------------------------
Mvan_0593|M.vanbaalenii_PYR-1 ---------MDDSSTYDVAIVGYGPVGATAANLLGRRGLKVVVIERDPDI
Mflv_2401|M.gilvum_PYR-GCK ---------MTEQTDVDVVIVGAGPAGLTLANILGLEGVRVLVVDERDKL
TH_0968|M.thermoresistible__bu ---------MTER--VDVVIVGAGPSGLTLANILGLQGVRTLVVEERDTL
MSMEG_4866|M.smegmatis_MC2_155 --------MMNSPGAVDVVIVGAGPVGLTLANILGGQGVRTLIIEERETL
MAB_2573c|M.abscessus_ATCC_199 -------MNASAQLSFPVVIIGAGPTGVTAATLLAQHGVQCLLLERWATV
MMAR_1503|M.marinum_M YGRARAISTDEEVMRIWQSVGLAERLQSDMLPDRPIAFVDAHGVPFIEMK
MAV_2518|M.avium_104 --------------------------------------------------
Mvan_0593|M.vanbaalenii_PYR-1 YFRARAISTDEEVMRIWQQIGLAEQLNAEMQPGCGADFVDADGRSFVKLL
Mflv_2401|M.gilvum_PYR-GCK IDYPRGVGLDDESLRTFQAIGLVDRVLPHTVPNQILRFFDAKRNLLAEMA
TH_0968|M.thermoresistible__bu IDYPRGVGLDDEALRTFQSIGLVDRILPHTVPNQILRFFDAKRRLLAEMA
MSMEG_4866|M.smegmatis_MC2_155 IDYPRGVGLDDESLRTFQSIGLVDAILPHTVPNQILRFYDANRRLLAEMA
MAB_2573c|M.abscessus_ATCC_199 YPQPRAVHLDDEIYRILAQLGIAEQFATISRPGAGMRLLDHRGQILSQIA
MMAR_1503|M.marinum_M IEPR-GSGHPPQQFLYQPAVDQVLRAGVARFANVEVLVEHECLRVLNNDA
MAV_2518|M.avium_104 --------------------------------------------------
Mvan_0593|M.vanbaalenii_PYR-1 PADR-GNGHPPQQFIYQPAVDKTLRAGVDRFPNVTVLLRHECLRLVPRTD
Mflv_2401|M.gilvum_PYR-GCK PPDA-RFGWPKRNGFVQPMVDAELFGGLQRFDNVEVRFGHRMHQCTQTTD
TH_0968|M.thermoresistible__bu PPDA-RFGWPKRNGFVQPMVDAELYAGLDRFDHVEVRFGHRMQNCVEASD
MSMEG_4866|M.smegmatis_MC2_155 PPDA-RFGWPKRNGFVQPMVDAELLAGLDRFDHVEVMWGRRMQTIAEDAD
MAB_2573c|M.abscessus_ATCC_199 RAPIGTHGYPQANMFDQPELEQLLRANLTNYPQVSLRGGVEVTTIAHISA
MMAR_1503|M.marinum_M A--AELLLADLRTDTFKRLRARYVIAADGGSSPTRGQLGVGYTGRTYAER
MAV_2518|M.avium_104 -----------------------MIAADGGSSPTRGQLGIGYSGRTYAER
Mvan_0593|M.vanbaalenii_PYR-1 D--VELMLADLSEDQFKRIRATYVIAADGGSSAVRGQLGIGFTGSTYSER
Mflv_2401|M.gilvum_PYR-GCK R----VEVEFDG--GQASVSARYVVGCDGGRSVTRRLMGVSFDGTTSSTR
TH_0968|M.thermoresistible__bu AGEDGVTVEFAD--GHPPVRASYVVGCDGGRSTTRRLMGVSFDGTTSSTR
MSMEG_4866|M.smegmatis_MC2_155 G----VTVEVSGPDGPASVHAQYVVGCDGGRSATRHLMGVSFDGTTSSTR
MAB_2573c|M.abscessus_ATCC_199 S-QVQVTFTDRDSGDEHKVTATYVLGCDGANSVVRAAIGASMTARRFAQR
::..**. * .* :* . . : *
MMAR_1503|M.marinum_M WVVIDTKVRRAWDAHDRLRFHCDPARPTVDCPTPLGHHRWEYPASADEAD
MAV_2518|M.avium_104 WVVIDTKVLKAWDAHDRLRFWANPARPTVDCPTPLGHHRWEYPARTDEDE
Mvan_0593|M.vanbaalenii_PYR-1 WIVIDTKVLREWPGHDRLRFHCNPARPTVDCPTPLGHHRWEFPVRDEEDE
Mflv_2401|M.gilvum_PYR-GCK WLVVD--IANDPLGHPNSEVGADPRRPYVSIAIAHGIRRFEFMIHPDETD
TH_0968|M.thermoresistible__bu WLVVD--VANDPLGHPNSEVGADPARPYVSISIAHGIRRFEFMIHPHETD
MSMEG_4866|M.smegmatis_MC2_155 WVVID--LANDPLGHPNSEVGADPQRPYASISIAHGIRRFEFMIHADETD
MAB_2573c|M.abscessus_ATCC_199 WLVVDLNTDADLGHWDGVHHLCDPDRATTFMRVGERRYRWEFKLHPGETA
*:*:* . .:* *. . *:*: *
MMAR_1503|M.marinum_M QKLVREEAIWRVLNAQG--ITDNEVEILRAVIYCHHVRVADRWRIGRVFL
MAV_2518|M.avium_104 RELLREEAIWQVLGDQG--ITAEHVEILRAVIYSHHVRVAERWRVGRIFL
Mvan_0593|M.vanbaalenii_PYR-1 RDLLTEDAIWKVLHGQG--ISKDDVEIIGFACYSHHVRFADRWRVGRIFL
Mflv_2401|M.gilvum_PYR-GCK EEADDPAFVRRMLGQLI--PYPERVDMIRHRVYTHHSRIAGSFREGRLML
TH_0968|M.thermoresistible__bu AEVDDPAFVRRMLGRRI--PYPERVDMIRYRVYTHHSRIAGSFRKGRLLL
MSMEG_4866|M.smegmatis_MC2_155 EQAEDPEFVAELLRPFV--PHPDRVDVIRRRVYTHHSRIAGSFRKGRMLL
MAB_2573c|M.abscessus_ATCC_199 ADYDTVEKLRPLIAVWTGASAISELTLIRVTEYTFRAHVASHWRDRNVFI
. : :: . : :: * .: :.* :* .:::
MMAR_1503|M.marinum_M AGDAAHAMPPWIGQGMSAGVRDAANLCWKLAAVLNGNAPDSLLDSYQDER
MAV_2518|M.avium_104 AGDAAHAMPPWIGQGMSAGVRDAANLCWKLAAVLRGQAPESLLDSYQTER
Mvan_0593|M.vanbaalenii_PYR-1 AGDAAHAMPPWIGQGMCAGVRDVGNLCWKLEAVLTGALSESVLDSYQAER
Mflv_2401|M.gilvum_PYR-GCK AGDAAHLMPVWQGQGYNSGIRDAANLGWKLAAVVTGRAGEELLDTYDVER
TH_0968|M.thermoresistible__bu AGDAAHLMPVWQGQGYNSGIRDAANLGWKLAAVVNGHAHDALLDTYDLER
MSMEG_4866|M.smegmatis_MC2_155 AGDAAHLMPVWQGQGYNSGIRDAFNLGWKLAAVVRGQAGDALLDTYDAER
MAB_2573c|M.abscessus_ATCC_199 LGDAAHLTPPFVGQGMGAGIRDAANLTWKLAGVLDGQLPTRVLDTYEQER
***** * : *** :*:**. ** *** .*: * :**:*: **
MMAR_1503|M.marinum_M KPHVTEVTRRAVFTGRLITERRRAVAAVRNGLGRALTRFPSAMARLQRAM
MAV_2518|M.avium_104 EPHVREVTRRACRVGRVITERNRAVVALRNHGLRALTRVPGVNAGLQKLT
Mvan_0593|M.vanbaalenii_PYR-1 LPHVREVTKRAVKVGRLIIERNPLRANIRNHFFRAASRLPFFTAWLRDHR
Mflv_2401|M.gilvum_PYR-GCK RKHARAMIDLSTMVGRVISPTNRKVAALRDRVIHAASAVPSLKRYVLEMR
TH_0968|M.thermoresistible__bu REHARAMIDLSTMVGRVISPTNRRVAALRDRLIHAASVVPTLKRYVLEMR
MSMEG_4866|M.smegmatis_MC2_155 RKHARAMIDLSTMVGRVISPTNRKVAALRDKLIRGASIVPTLKRYVLEMR
MAB_2573c|M.abscessus_ATCC_199 KPHAWKMITIALAVGAAMTARGTAATAIRNLLVPRLQYIPGLRSRIQHSG
*. : : .* : . :*: .* :
MMAR_1503|M.marinum_M WIPPARYQDGFLARG-----HRAVGWQIPQPWVIDGNGATARLDDILGSQ
MAV_2518|M.avium_104 WIPGARYRDGFFAAGK----TRAVGWQIPQPWVAEPNGARIRLDDLLGGQ
Mvan_0593|M.vanbaalenii_PYR-1 WLPDARYRAGLLTSDR----HPAVGWLIPQPWVVDEKGDRVRLDDVLAGG
Mflv_2401|M.gilvum_PYR-GCK FKPMPRYQQGAVLHHAHAAPNSPTGTLFIQPRVDTRDTQNALLDDVLGTG
TH_0968|M.thermoresistible__bu FKPMPRYLQGAVYHGGPRSKSSPTGTLFIQPRVDTRDAENVLLDDVLGPG
MSMEG_4866|M.smegmatis_MC2_155 FKPMPRYHEGAVYHAKPPTPASPVGTLFIQPRVDTREQDNVLLDDVLGTG
MAB_2573c|M.abscessus_ATCC_199 TPALTRSALVNKTPRCR----QLAGTQIPNLVLSQG----HRIDTLLGSG
. .* .* : : : :* :*.
MMAR_1503|M.marinum_M WVLLHTGDAP------TGSQAWTQRGVPAVRLGPPESGARPDG-------
MAV_2518|M.avium_104 WAILHIGPAP------AGAQEWTRLDVPLIQIT-----------------
Mvan_0593|M.vanbaalenii_PYR-1 WTVLHIGPE-------SLWRSWRSAGVPVIAIAPPGSAPAPGR-------
Mflv_2401|M.gilvum_PYR-GCK FAVVCWSNNLRAVLGEEAFGRWKALGAKFVEVRPMSQLRWPGHDDPDVAV
TH_0968|M.thermoresistible__bu FAVLCWSNNLRAVLGEDAFSHWKSLGATFVEARPITQLHWPGHDDPDVVV
MSMEG_4866|M.smegmatis_MC2_155 FAVLCWNNNPRTLLGEAAFTRWKALGATFIAARPSTQLHWTKDDDPDVVI
MAB_2573c|M.abscessus_ATCC_199 FALVASETVD-----ADQQAQLRRRGAAIIVEP-----------------
:.:: .. :
MMAR_1503|M.marinum_M IRDSGGTLLRWLARKHASAVVLRPDGFVYAAAESGSPLPPPP--------
MAV_2518|M.avium_104 ----DAPLIRWLGHKKAAAVVLRPDGFIYAAAQSGHRLPKPP--------
Mvan_0593|M.vanbaalenii_PYR-1 IVDREGTLIRWIQTKKATVVVVRPDGFVYAAAAD-QPLPAPP--------
Mflv_2401|M.gilvum_PYR-GCK IGDRTGALKAWFDVHTESVLFVRPDRCIAAACIAQRAPEISEGLFAALHL
TH_0968|M.thermoresistible__bu VGDRTGALKGWFDVHTDSVLFLRPDRCIAAACIAQRAPETSAALSEVMCL
MSMEG_4866|M.smegmatis_MC2_155 VGDRTGALKAFFDAHTESVLVLRPDRCIAGADIAQRAPELSTALFGILHL
MAB_2573c|M.abscessus_ATCC_199 ---RGSELDRWLRRGGARAALIRPDRTVMQTARSPSQIAFTT--------
. * :: . .:*** : : .
MMAR_1503|M.marinum_M --------------------AGHFGNTLPTSMKIEALA-
MAV_2518|M.avium_104 --------------------PGYSGNTITTPSKTEVSA-
Mvan_0593|M.vanbaalenii_PYR-1 --------------------AGFTPRVRQQA--------
Mflv_2401|M.gilvum_PYR-GCK TSGAHLTQGGGTDGEKPDRAVLHVAQPAAESSGTGAGTS
TH_0968|M.thermoresistible__bu T------QGGGNGAASP---VLHVAQSAAEPSGTATRST
MSMEG_4866|M.smegmatis_MC2_155 R------QGGENGATGP---VLYVPQPTAESSGTVGRAS
MAB_2573c|M.abscessus_ATCC_199 --------------------TLFAPTHQPAPTYRETP--
. .