For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SRRDDVQGRPAVDTRIPESMAARLQRAQSWAIGSDPGLLRLRMATRTTAALGCSLLVLYLLTMATGQPLT VALIGVVITMIAARSVDDPDSRQQQITMALLPVPAALSISVAALLAPHLIAAEVAFVVIVFAAVYVRRFG PRGRALGMVAFMAYFFSLYLRAGVAQLPWMIGAVLVGTLCTFVMSTYVLPDRPESVLRATIRSLRARMAI VVDTAADTVRAGRLDERRRRRMRARIARLNETALLVQSQIEDKTHPATLWPGISAEQLAPWLFDAELSVE WVAIAGHRAMAIAEEIPAATRIELADTLTQLAWAIRGPRTEGLHQAASRAQRLLDGQPAPAPTDDPSQGA VRRLGLAIVAAATATSEVRELIGDATAAPPDPDNNDVVQPVPEDTAADGGPGEAGLAPTTRQAIQVSIAA SLAIVTGELVSPLHWYWAVIAAFVVFAGTNSWGETLTKGWQRLLGTVLGVPCGVLVATVVAGDKVFSVVM IFACLFFAVYFMQVTYSLMTFWITTMLALLYGLLGKFTFGVLMLRIEETAIGAVIGVAVAILVLPTTIGT VVRRDTRAFLTTLSELIEISIATMFDDDVTDSPTERARELDRDLQQFRVTAKPMLAGVAGFAGRRSIQHS VRLLTICDRYARILARRSEQYLDPGCPHGLSDAITTAAAQIRRNIDALVTALDGTQDVAITPAADSLDAA EAVVRQHADALDRADPGSNPRHLLAAVNSLRRIEGAVVNAAVHFGADADLMAHDQPG*
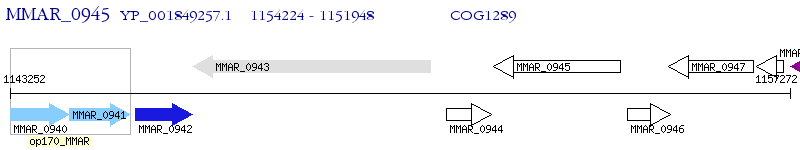
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_0945 | - | - | 100% (758) | hypothetical protein MMAR_0945 |
| M. marinum M | MMAR_1191 | sugI | 5e-05 | 26.99% (163) | sugar-transport integral membrane protein SugI |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0746 | - | 6e-12 | 22.28% (561) | membrane protein-like protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_02288 | - | 3e-12 | 53.23% (62) | hypothetical protein MLBr_02288 |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. avium 104 | MAV_4566 | - | 0.0 | 65.84% (726) | hypothetical protein MAV_4566 |
| M. smegmatis MC2 155 | MSMEG_0090 | - | 1e-11 | 21.83% (623) | hypothetical protein MSMEG_0090 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_1444 | sugI | 1e-05 | 27.61% (163) | sugar-transport integral membrane protein SugI |
| M. vanbaalenii PYR-1 | Mvan_0100 | - | 1e-13 | 21.76% (703) | membrane protein-like protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0746|M.gilvum_PYR-GCK --------------------MLSLRQRVADRVAQRDPDRDALRRAVRAAV
Mvan_0100|M.vanbaalenii_PYR-1 ----------------MAAAPHELRHRLVRRVAQRDPDNDALRRALRAAA
MSMEG_0090|M.smegmatis_MC2_155 ----------------MLLTPAKLWRRAAERLRERDPEYDALRRAARAGV
MMAR_0945|M.marinum_M MSRRDDVQGRPAVDTRIPESMAARLQRAQSWAIGSDPGLLRLRMATRTTA
MAV_4566|M.avium_104 -------------------------------MIGSDPGLLRLRMATRTTA
MLBr_02288|M.leprae_Br4923 --------------------------------------------------
MUL_1444|M.ulcerans_Agy99 --------------------------MQRGILVALTAASVGLIYGYDLSI
Mflv_0746|M.gilvum_PYR-GCK VVPAAGAVSFAVAGGSTQTPLFTIFGAVALLVFSDFPGNRQNR--AVAYG
Mvan_0100|M.vanbaalenii_PYR-1 VVPLAAALSFALAGGSAQTPLFTIFGSTALLVFSDFPGNRQNR--AVAYA
MSMEG_0090|M.smegmatis_MC2_155 VLPIAAAIGFAVGGGS-QTPLFSIFGAVSLLITADFPGNRPAR--ALAFA
MMAR_0945|M.marinum_M ALGCSLLVLYLLTMATGQPLTVALIGVVITMIAARSVDDPDSRQQQITMA
MAV_4566|M.avium_104 ALGLSLLALWLLTRATGQPLTVALLGVVITMVASRSVNEPDPRQQRITMA
MLBr_02288|M.leprae_Br4923 --------------------------------------------------
MUL_1444|M.ulcerans_Agy99 IAGAQLFITEEFGLSTHQQELLTTMVVIGQIVG------------ALGAG
Mflv_0746|M.gilvum_PYR-GCK GLGLNGFLLITLGTLVAPYPWPAVAAMFVIGVAVTFSGVLSETLAAGQRA
Mvan_0100|M.vanbaalenii_PYR-1 GLGFNGFVLITLGTLVAPHPWPAVAVMFALGVVVTFSGVLSETIAAGQRA
MSMEG_0090|M.smegmatis_MC2_155 GLAFNGAVLIVLGTLIAPHVWLSVVSMFVIGVVVSFSGVLSEVVAAGQRA
MMAR_0945|M.marinum_M LLPVPAALSISVAALLAPHLIAAEVAFVVIVFAAVYVRRFGPRGRALGMV
MAV_4566|M.avium_104 LLPVPAAVAITAAAVLAPHPVAGDVVFVLIVFAAAYIRRFGARGRALGMV
MLBr_02288|M.leprae_Br4923 --------------------------------------------------
MUL_1444|M.ulcerans_Agy99 VLANAIGRKKSVVMLLVAYTMFAVLGALSVSLPMLLAARFLLGLAVGVSI
Mflv_0746|M.gilvum_PYR-GCK TLLTFVLPACTPPGPVAERLLG--WTIALAVCVPAALFLLPPRHHGQLRR
Mvan_0100|M.vanbaalenii_PYR-1 TLLTFVLPACTPAGPVGERLLG--WAIALAVCVPAALFVLPPRHHGQLRR
MSMEG_0090|M.smegmatis_MC2_155 TLLLFVLPLCTPVGPIADRLLG--WVIALVVCVPAALFLFPPRHHDELRR
MMAR_0945|M.marinum_M AFMAYFFSLYLRAGVAQLPWMIGAVLVGTLCTFVMSTYVLPDRPESVLRA
MAV_4566|M.avium_104 AFMSYFFTLYLRARVSELPWMIGAVAVGTVCTYVMTSHVLPDRPERVLRA
MLBr_02288|M.leprae_Br4923 --------------------------------------------------
MUL_1444|M.ulcerans_Agy99 VVVPVYVAESAPAAVRGS------LLTVYQLTTVSGLIVGYLTGYLLAGT
Mflv_0746|M.gilvum_PYR-GCK RAARACTALADRLDGTVTPDDVQRAMDALRKTFLGADFRPVGLTAGSRAL
Mvan_0100|M.vanbaalenii_PYR-1 RAARACRALADRLDGSATPADVQRAMDALRATFLGADFRPVGLTAGSRAL
MSMEG_0090|M.smegmatis_MC2_155 TAARVCRVLAYRLEGSASARDVNRAMNELYESFLGADYRPVALTAGSRAL
MMAR_0945|M.marinum_M TIRSLRARMAIVVDTAADTVRAGRLDERRRRRMRARIARLNETALLVQSQ
MAV_4566|M.avium_104 TIRALRARMAIVIDTTAEAVRTGRLDERRRRRMRARTVRLNETALMVQSQ
MLBr_02288|M.leprae_Br4923 --------------------------------------------------
MUL_1444|M.ulcerans_Agy99 HSWRWMLGLATVPAMLLLPLLIRMPDTPRWYVMKGRIQEARAALLRVDPA
Mflv_0746|M.gilvum_PYR-GCK VRVVDDLEWVADRTGRGTGVTLRDK-DAVVRVLRASAAVLSISRPADREM
Mvan_0100|M.vanbaalenii_PYR-1 VRVVDDLEWVADRVGRNTGVALRDK-DAVVRVLRCAAAVLQISRPADRDA
MSMEG_0090|M.smegmatis_MC2_155 VRVVDDLGWISDRVTDSTGEILGEMRDPVVRVLRDSAALLRIHDPARRAE
MMAR_0945|M.marinum_M IEDKTHPATLWPGISAEQLAPWLFDAELSVEWVAIAGHRAMAIAEEIPAA
MAV_4566|M.avium_104 IEDRADPGTLWPGVTAEQLAPWLLDAELAIEWVATAGRRAAALGAELPEA
MLBr_02288|M.leprae_Br4923 --------------------------------------------------
MUL_1444|M.ulcerans_Agy99 ADVEEELAEIGTALSEGSGG--------------------------VSEM
Mflv_0746|M.gilvum_PYR-GCK ARAELEGALLALRTLARGRWREDIDEILG-AADDEQAVAQGRDLLRRRTI
Mvan_0100|M.vanbaalenii_PYR-1 ARADLEAALITLRTVARGRWRDDLDAILS-APDDAQAVDLGRDLLRRRTI
MSMEG_0090|M.smegmatis_MC2_155 RSADLRAALEELRTVSQGTYREGIAAILG-TPDDADAVEVGKALLVRRTI
MMAR_0945|M.marinum_M TRIELADTLTQLAWAIRGPRTEGLHQAASRAQRLLDGQPAPAPTDDPSQG
MAV_4566|M.avium_104 VRAELVSALTQLGRAIRLPEPGGLRDAASRARRLLD----AATDDRPAGT
MLBr_02288|M.leprae_Br4923 --------------------------------------------------
MUL_1444|M.ulcerans_Agy99 LRPPFLRATIFVITLGFLIQITGINAIIYYSPRIFEAMGFTGDFALLG--
Mflv_0746|M.gilvum_PYR-GCK AATVSATGRVIAAAAAADARPVWARALGLRLPPTGASDRLLPETVAAAKI
Mvan_0100|M.vanbaalenii_PYR-1 AATIGATGRVISAAAAADARPVWARALGLRLPPTGASDRLLPETVAAAQI
MSMEG_0090|M.smegmatis_MC2_155 AATVAVTGRVIRNAALADARPVWARVLGRRLPETGSADWIMPETTAVAEI
MMAR_0945|M.marinum_M AVRRLGLAIVAAATATSEVRELIGDAT-AAPPDPDNND----VVQPVPED
MAV_4566|M.avium_104 EVRRLALAIINAATATADVRAIVDGAAGAAVPDVAGSERPPAAAGVAAEE
MLBr_02288|M.leprae_Br4923 --------------------------------------------------
MUL_1444|M.ulcerans_Agy99 ---LPALVQIAGVAAVITSLLLVDRVGRRPILLSGIAIMIVADVALMAVF
Mflv_0746|M.gilvum_PYR-GCK TTG----FVENRSVAVRNSLRTGLGLALAVTVTHVFPVEHGFWVVLGALS
Mvan_0100|M.vanbaalenii_PYR-1 TSG----FLANRSVAVRNSLRTGLGLALAVAVTHLFPVEHGFWVVLGALS
MSMEG_0090|M.smegmatis_MC2_155 AKG----FLSTRAVVLRNSLRTGLGLALAVAVTHVFPVEHGFWVVLGAMS
MMAR_0945|M.marinum_M TAADGGPGEAGLAPTTRQAIQVSIAASLAIVTGELVSPLHWYWAVIAAFV
MAV_4566|M.avium_104 PAEQEEEQPAGLRPTTRQAIQVSVAASLAIITGELVSPARWYWAVIAAFV
MLBr_02288|M.leprae_Br4923 --------------------------------------------MVAAFV
MUL_1444|M.ulcerans_Agy99 ARG-------------QGAAILGFAGILLFIIGYTMGFGSLGWVYASESF
.
Mflv_0746|M.gilvum_PYR-GCK VLR--SSALTTGTRVWRAVVGTGIGFLLGALLISLVGVDPVVMWILMPLA
Mvan_0100|M.vanbaalenii_PYR-1 VLR--SSALTTGTRVWRAVFGTAIGFLFGAVLISLVGVNPVLLWLLMPVV
MSMEG_0090|M.smegmatis_MC2_155 VLR--SSALTTGTRVVRAVAGTALGFVIGAVLIELVGVQPVVLWILLPIV
MMAR_0945|M.marinum_M VFAGTNSWGETLTKGWQRLLGTVLGVPCGVLVATVVAGDKVFSVVMIFAC
MAV_4566|M.avium_104 IFAGTNSWGETLTKGWQRLLGTMLGVPAGVLVATLLTGHEAAALAGIFVC
MLBr_02288|M.leprae_Br4923 ILAVTNSWGETLTRGWQQLLVNTVG----VLVTTLFSGNTTVSPAAIFVC
MUL_1444|M.ulcerans_Agy99 PTR-LRSIGSSTMLTANLIANAIVAAVFLTMLHSLGGSGAFAVFGVLALV
* : . : :. .:: : :
Mflv_0746|M.gilvum_PYR-GCK VFGSAYVPEIASFIAAQAAFTMMVLISFNLIAPTGWEVGLIRVEDVFVGA
Mvan_0100|M.vanbaalenii_PYR-1 VFGSAYVPEIASFTAAQAAFTMMVLIFFNLIAPTGWQVGLIRVEDVVMGT
MSMEG_0090|M.smegmatis_MC2_155 AFGSAYVPEVASFIAGQAAFTMMVLINFNLIAPTGWQVGLIRIEDVVVGA
MMAR_0945|M.marinum_M LFFAVYFMQVTYSLMTFWITTMLALL-YGLLGKFTFGVLMLRIEETAIGA
MAV_4566|M.avium_104 LFCAFYFMTVTYSLMTFWITTMLALL-YGLLGQFSFGVLMLRIEETALGA
MLBr_02288|M.leprae_Br4923 LFCAFYIYDGN-----------------------------LQPDARCMGN
MUL_1444|M.ulcerans_Agy99 AFGVVYRYAPET-------------------------------KGRQLEE
* * . :
Mflv_0746|M.gilvum_PYR-GCK LVGVVVSVLLWPRGATASVTRAIDSARFVAARYLRAAVLRITRGAYEERT
Mvan_0100|M.vanbaalenii_PYR-1 LVAAAVSMLLWPRGATASVTRAIDSARTVFATYLRTAVLRITRGASEART
MSMEG_0090|M.smegmatis_MC2_155 LVGIVVSILLWPRGARASVTKAIDAARAAGAELLEAAVLRVTRGASEEAT
MMAR_0945|M.marinum_M VIGVAVAILVLPTTIGTVVRRDTRAFLTTLSELIEISIATMFDDDVTDSP
MAV_4566|M.avium_104 VIGATVAVVVLPTNTRTAIRADTRAFLTSLSALIEACAAAMSGG--AASP
MLBr_02288|M.leprae_Br4923 T-------------------------------------------------
MUL_1444|M.ulcerans_Agy99 IRHFWENGGRWPEKLTEAP-------------------------------
Mflv_0746|M.gilvum_PYR-GCK NEVVTLGHAALDASRVVDDAVRHYLSESSGETDFR-------------AP
Mvan_0100|M.vanbaalenii_PYR-1 DEVATRSHAALAASRVIDDAVRQYLSESSGETDFR-------------AP
MSMEG_0090|M.smegmatis_MC2_155 DRVIALSHSALEASRTLDDTVRHYLSESGGPSDQR-------------AP
MMAR_0945|M.marinum_M TERARELDRDLQQFRVTAKPMLAGVAGFAGRRSIQHSVRLLTICDRYARI
MAV_4566|M.avium_104 SEQARQLDRDLQQFRVTAKPLLAGVAGLAGRRGIRRALRIFTACDRYGRA
MLBr_02288|M.leprae_Br4923 --------------------------------------------------
MUL_1444|M.ulcerans_Agy99 --------------------------------------------------
Mflv_0746|M.gilvum_PYR-GCK VVRSSNRATRLRGAADLIADIPTPPPLGTYPKVRSVLEAHADAICARVSG
Mvan_0100|M.vanbaalenii_PYR-1 IVRSFNRAIRLRGAADLVADIPTPPPLGTYPSVRAVVESHADAICERVAG
MSMEG_0090|M.smegmatis_MC2_155 IVRSATRAIRVRAAAELIADV-VPPPLEAYQETRKTIEAHARIICARLTG
MMAR_0945|M.marinum_M LARRSEQYLD-PGCPHGLSDAITTAAAQIRRNIDALVTALDGTQDVAITP
MAV_4566|M.avium_104 LARSSEQYRGSPGPGPQLAQAFSAAAAQTRRNLDALLEAIDSGHPPTLVS
MLBr_02288|M.leprae_Br4923 --------------------------------------------------
MUL_1444|M.ulcerans_Agy99 --------------------------------------------------
Mflv_0746|M.gilvum_PYR-GCK TSEPVGAWAPISEDFVRALRSEAPEDT----LGVAAALPLLTAAAALGEL
Mvan_0100|M.vanbaalenii_PYR-1 RADPDHSWEPISDDFVRALRSEARGDD----LGLSAALPLLTAAAALGEL
MSMEG_0090|M.smegmatis_MC2_155 -EDTTSVFVPIGETFVLALRAEATGGD----LAVSAALPLVTVAAHIGEL
MMAR_0945|M.marinum_M AADSLDAAEAVVRQHADALDRADPGSNPRHLLAAVNSLRRIEGAVVNAAV
MAV_4566|M.avium_104 AADELDAAETAARQHDSDGD-GETRPDVRRFLTAVHALRQIERAVISTAT
MLBr_02288|M.leprae_Br4923 --------------------------------------------------
MUL_1444|M.ulcerans_Agy99 --------------------------------------------------
Mflv_0746|M.gilvum_PYR-GCK ELIYPWDTGPS-----
Mvan_0100|M.vanbaalenii_PYR-1 ELIYPRPATG------
MSMEG_0090|M.smegmatis_MC2_155 ELLYPQPVESVH----
MMAR_0945|M.marinum_M HFGADADLMAHDQPG-
MAV_4566|M.avium_104 NLGGREDVRTATAAPR
MLBr_02288|M.leprae_Br4923 ----------------
MUL_1444|M.ulcerans_Agy99 ----------------