For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VIGSDPGLLRLRMATRTTAALGLSLLALWLLTRATGQPLTVALLGVVITMVASRSVNEPDPRQQRITMAL LPVPAAVAITAAAVLAPHPVAGDVVFVLIVFAAAYIRRFGARGRALGMVAFMSYFFTLYLRARVSELPWM IGAVAVGTVCTYVMTSHVLPDRPERVLRATIRALRARMAIVIDTTAEAVRTGRLDERRRRRMRARTVRLN ETALMVQSQIEDRADPGTLWPGVTAEQLAPWLLDAELAIEWVATAGRRAAALGAELPEAVRAELVSALTQ LGRAIRLPEPGGLRDAASRARRLLDAATDDRPAGTEVRRLALAIINAATATADVRAIVDGAAGAAVPDVA GSERPPAAAGVAAEEPAEQEEEQPAGLRPTTRQAIQVSVAASLAIITGELVSPARWYWAVIAAFVIFAGT NSWGETLTKGWQRLLGTMLGVPAGVLVATLLTGHEAAALAGIFVCLFCAFYFMTVTYSLMTFWITTMLAL LYGLLGQFSFGVLMLRIEETALGAVIGATVAVVVLPTNTRTAIRADTRAFLTSLSALIEACAAAMSGGAA SPSEQARQLDRDLQQFRVTAKPLLAGVAGLAGRRGIRRALRIFTACDRYGRALARSSEQYRGSPGPGPQL AQAFSAAAAQTRRNLDALLEAIDSGHPPTLVSAADELDAAETAARQHDSDGDGETRPDVRRFLTAVHALR QIERAVISTATNLGGREDVRTATAAPR
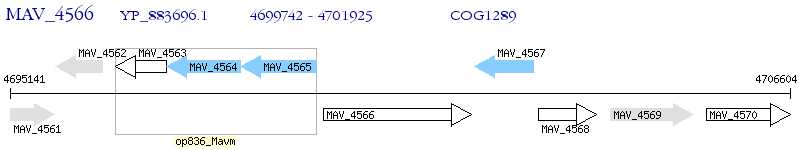
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_4566 | - | - | 100% (727) | hypothetical protein MAV_4566 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0746 | - | 4e-12 | 23.67% (562) | membrane protein-like protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_02288 | - | 2e-15 | 61.29% (62) | hypothetical protein MLBr_02288 |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_0945 | - | 0.0 | 65.84% (726) | hypothetical protein MMAR_0945 |
| M. smegmatis MC2 155 | MSMEG_5547 | - | 3e-11 | 25.18% (413) | hypothetical protein MSMEG_5547 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_0100 | - | 2e-13 | 22.68% (732) | membrane protein-like protein |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_4566|M.avium_104 -------------------------------MIGSDPG--LLRLRMATRT
MMAR_0945|M.marinum_M MSRRDDVQGRPAVDTRIPESMAARLQRAQSWAIGSDPG--LLRLRMATRT
MLBr_02288|M.leprae_Br4923 --------------------------------------------------
Mflv_0746|M.gilvum_PYR-GCK --------------------MLSLRQRVADRVAQRDPD--RDALRRAVRA
Mvan_0100|M.vanbaalenii_PYR-1 ----------------MAAAPHELRHRLVRRVAQRDPD--NDALRRALRA
MSMEG_5547|M.smegmatis_MC2_155 -------------------MLVTMSERHAPEPVGNPLRGIFVVNRVAGRW
MAV_4566|M.avium_104 TAALGLSLLALWLLTRATGQPLTVALLGVVITMVASRSVNEPDPRQQRIT
MMAR_0945|M.marinum_M TAALGCSLLVLYLLTMATGQPLTVALIGVVITMIAARSVDDPDSRQQQIT
MLBr_02288|M.leprae_Br4923 --------------------------------------------------
Mflv_0746|M.gilvum_PYR-GCK AVVVPAAGAVSFAVAGGSTQTPLFTIFGAVALLVFSDFPGNRQNR--AVA
Mvan_0100|M.vanbaalenii_PYR-1 AAVVPLAAALSFALAGGSAQTPLFTIFGSTALLVFSDFPGNRQNR--AVA
MSMEG_5547|M.smegmatis_MC2_155 PFALRAAICMAVPVLVGWSAGNTSAGLIATIGGFTSLYGSGRPYLNRGVF
MAV_4566|M.avium_104 MALLPVPAAVAITAAAVLAPHPVAGDVVFVLIVFAAAYIRRFGARGRALG
MMAR_0945|M.marinum_M MALLPVPAALSISVAALLAPHLIAAEVAFVVIVFAAVYVRRFGPRGRALG
MLBr_02288|M.leprae_Br4923 --------------------------------------------------
Mflv_0746|M.gilvum_PYR-GCK YGGLGLNGFLLITLGTLVAPYPWPAVAAMFVIGVAVTFSGVLSETLAAGQ
Mvan_0100|M.vanbaalenii_PYR-1 YAGLGFNGFVLITLGTLVAPHPWPAVAVMFALGVVVTFSGVLSETIAAGQ
MSMEG_5547|M.smegmatis_MC2_155 LGGVAVTFAVVVALGDWASRHVWLGVATVTVIAMVTVLVCHALSVGPPGA
MAV_4566|M.avium_104 MVAFMSYFFTLYLR-ARVSELPWMIGAVAVGTVCTYVMTSHVLPDRPERV
MMAR_0945|M.marinum_M MVAFMAYFFSLYLR-AGVAQLPWMIGAVLVGTLCTFVMSTYVLPDRPESV
MLBr_02288|M.leprae_Br4923 --------------------------------------------------
Mflv_0746|M.gilvum_PYR-GCK RATLLTFVLPACTP-PGPVAERLLG--WTIALAVCVPAALFLLPPRHHGQ
Mvan_0100|M.vanbaalenii_PYR-1 RATLLTFVLPACTP-AGPVGERLLG--WAIALAVCVPAALFVLPPRHHGQ
MSMEG_5547|M.smegmatis_MC2_155 YMFVLACAAGTGAPGTADLGPVRMALLVLAGGSFAWCVHMLGALIRPRGP
MAV_4566|M.avium_104 LRATIRALRARMAIVIDTTAEAVRTGRLDERRRRRMRARTVRLNETALMV
MMAR_0945|M.marinum_M LRATIRSLRARMAIVVDTAADTVRAGRLDERRRRRMRARIARLNETALLV
MLBr_02288|M.leprae_Br4923 --------------------------------------------------
Mflv_0746|M.gilvum_PYR-GCK LRRRAARACTALADRLDGTVTPDDVQRAMDALRKTFLG--ADFRPVGLTA
Mvan_0100|M.vanbaalenii_PYR-1 LRRRAARACRALADRLDGSATPADVQRAMDALRATFLG--ADFRPVGLTA
MSMEG_5547|M.smegmatis_MC2_155 ERAAVRDAALAIAEYIDSIGGDANAEG-----------------------
MAV_4566|M.avium_104 QSQIEDRADPGTLWPGVTAEQLAPWLLDAELAIEWVATAGRRAAALGAEL
MMAR_0945|M.marinum_M QSQIEDKTHPATLWPGISAEQLAPWLFDAELSVEWVAIAGHRAMAIAEEI
MLBr_02288|M.leprae_Br4923 --------------------------------------------------
Mflv_0746|M.gilvum_PYR-GCK GSRALVRVVDDLEWVADRTGRGTGVTLRDKDAVVRVLRASAAVLSISRPA
Mvan_0100|M.vanbaalenii_PYR-1 GSRALVRVVDDLEWVADRVGRNTGVALRDKDAVVRVLRCAAAVLQISRPA
MSMEG_5547|M.smegmatis_MC2_155 QARHRAATLLHTAWVTLVTFQPVQPKPDQALYRLRVVNRRLHVLLADAMR
MAV_4566|M.avium_104 P-EAVRAELVSALTQLGRAIRLPEPGGLRDAASRARRLLD----AATDDR
MMAR_0945|M.marinum_M P-AATRIELADTLTQLAWAIRGPRTEGLHQAASRAQRLLDGQPAPAPTDD
MLBr_02288|M.leprae_Br4923 --------------------------------------------------
Mflv_0746|M.gilvum_PYR-GCK DREMARAELEGALLALRTLARGRWREDIDEILGAADDEQAVAQGRDLLRR
Mvan_0100|M.vanbaalenii_PYR-1 DRDAARADLEAALITLRTVARGRWRDDLDAILSAPDDAQAVDLGRDLLRR
MSMEG_5547|M.smegmatis_MC2_155 AADAGTAPPPGGAAFARELAAIPPLTDEHGAEGVP------------LGR
MAV_4566|M.avium_104 PAGTEVRRLALAIINAATATADVRAIVDGAAGAAVPDVAGSERPPAAAGV
MMAR_0945|M.marinum_M PSQGAVRRLGLAIVAAATATSEVRELIGDAT-AAPPDPDNND----VVQP
MLBr_02288|M.leprae_Br4923 --------------------------------------------------
Mflv_0746|M.gilvum_PYR-GCK RTIAATVSATGRVIAAAAAADARPVWARALGLRLPPTGASDR-----LLP
Mvan_0100|M.vanbaalenii_PYR-1 RTIAATIGATGRVISAAAAADARPVWARALGLRLPPTGASDR-----LLP
MSMEG_5547|M.smegmatis_MC2_155 PGAGQLLRQALR--------------------------------------
MAV_4566|M.avium_104 AAEEPAEQEEEQPAGLRPTTRQAIQVSVAASLAIITGELVSPARWYWAVI
MMAR_0945|M.marinum_M VPEDTAADGGPGEAGLAPTTRQAIQVSIAASLAIVTGELVSPLHWYWAVI
MLBr_02288|M.leprae_Br4923 ------------------------------------------------MV
Mflv_0746|M.gilvum_PYR-GCK ETVAAAKITTGFVENRSVAVRNSLRTGLGLALAVTVTHVFPVEHGFWVVL
Mvan_0100|M.vanbaalenii_PYR-1 ETVAAAQITSGFLANRSVAVRNSLRTGLGLALAVAVTHLFPVEHGFWVVL
MSMEG_5547|M.smegmatis_MC2_155 ----------PGSMSLRLAARVAVAVAISGAIAVGFAEALAIDRAYWAMA
:
MAV_4566|M.avium_104 AAFVIFAGTNSWGETLTKGWQRLLGTMLGVPAGVLVATLLTGHEAAALAG
MMAR_0945|M.marinum_M AAFVVFAGTNSWGETLTKGWQRLLGTVLGVPCGVLVATVVAGDKVFSVVM
MLBr_02288|M.leprae_Br4923 AAFVILAVTNSWGETLTRGWQQLLVNTVG----VLVTTLFSGNTTVSPAA
Mflv_0746|M.gilvum_PYR-GCK GALSVLR--SSALTTGTRVWRAVVGTGIGFLLGALLISLVGVDPVVMWIL
Mvan_0100|M.vanbaalenii_PYR-1 GALSVLR--SSALTTGTRVWRAVFGTAIGFLFGAVLISLVGVNPVLLWLL
MSMEG_5547|M.smegmatis_MC2_155 AAVLMLHQGFAWVRTVTRSVERMVGTWLGLGVAGAILAVHPQGVWLALTI
.*. :: : * *: . :. . :* : ::
MAV_4566|M.avium_104 IFVCLFCAFYFMTVTYSLMTFWITTMLALLYG-LLGQFSFGVLMLRIEET
MMAR_0945|M.marinum_M IFACLFFAVYFMQVTYSLMTFWITTMLALLYG-LLGKFTFGVLMLRIEET
MLBr_02288|M.leprae_Br4923 IFVCLFCAFYIYDGN-----------------------------LQPDAR
Mflv_0746|M.gilvum_PYR-GCK MPLAVFGSAYVPEIASFIAAQAAFTMMVLISFNLIAPTGWEVGLIRVEDV
Mvan_0100|M.vanbaalenii_PYR-1 MPVVVFGSAYVPEIASFTAAQAAFTMMVLIFFNLIAPTGWQVGLIRVEDV
MSMEG_5547|M.smegmatis_MC2_155 GVLQFVIEMYVVRNYTLAVVFITPAALTISSG----GAAITDLGALLWAR
.. *.
MAV_4566|M.avium_104 ALGAVIGATVAVVVLPTNTRTAIRADTRAFLTSLSALIEACAAAMSGG--
MMAR_0945|M.marinum_M AIGAVIGVAVAILVLPTTIGTVVRRDTRAFLTTLSELIEISIATMFDDDV
MLBr_02288|M.leprae_Br4923 CMGNT---------------------------------------------
Mflv_0746|M.gilvum_PYR-GCK FVGALVGVVVSVLLWPRGATASVTRAIDSARFVAARYLRAAVLRITRGAY
Mvan_0100|M.vanbaalenii_PYR-1 VMGTLVAAAVSMLLWPRGATASVTRAIDSARTVFATYLRTAVLRITRGAS
MSMEG_5547|M.smegmatis_MC2_155 GVDTLIGCAVALLAYRATQRMRAPTALSTAITRTLDAVDATLPHLGAGAA
:.
MAV_4566|M.avium_104 AASPSEQARQLDRDLQQFRVTAKPLLAGVAGLAGRRGIRRALRIFTACDR
MMAR_0945|M.marinum_M TDSPTERARELDRDLQQFRVTAKPMLAGVAGFAGRRSIQHSVRLLTICDR
MLBr_02288|M.leprae_Br4923 --------------------------------------------------
Mflv_0746|M.gilvum_PYR-GCK EERTNEVVTLGHAALDASRVVDDAVRHYLSESSGETDFR-----------
Mvan_0100|M.vanbaalenii_PYR-1 EARTDEVATRSHAALAASRVIDDAVRQYLSESSGETDFR-----------
MSMEG_5547|M.smegmatis_MC2_155 TTGAARTHRR------DLHVRAMALLPSYDAAVGG---------------
MAV_4566|M.avium_104 YGRALARSSEQYRGSPGPGPQLAQAFSAAAAQTRRNLDALLEAIDSGHPP
MMAR_0945|M.marinum_M YARILARRSEQYLD-PGCPHGLSDAITTAAAQIRRNIDALVTALDGTQDV
MLBr_02288|M.leprae_Br4923 --------------------------------------------------
Mflv_0746|M.gilvum_PYR-GCK --APVVRSSNRATRLRGAADLIADIPTPPPLGTYPKVRSVLEAHADAICA
Mvan_0100|M.vanbaalenii_PYR-1 --APIVRSFNRAIRLRGAADLVADIPTPPPLGTYPSVRAVVESHADAICE
MSMEG_5547|M.smegmatis_MC2_155 -----------SAAQRRAAERLWPTVVAAEQLAYRTLAACWALERDGDVD
MAV_4566|M.avium_104 TLVSAADELDAAETAARQHDSDGD-GETRPDVRRFLTAVHALRQIERAVI
MMAR_0945|M.marinum_M AITPAADSLDAAEAVVRQHADALDRADPGSNPRHLLAAVNSLRRIEGAVV
MLBr_02288|M.leprae_Br4923 --------------------------------------------------
Mflv_0746|M.gilvum_PYR-GCK RVSGTSEPVGAWAPISEDFVRALRSEAPEDTLG---------VAAALPLL
Mvan_0100|M.vanbaalenii_PYR-1 RVAGRADPDHSWEPISDDFVRALRSEARGDDLG---------LSAALPLL
MSMEG_5547|M.smegmatis_MC2_155 AARDTADALAEQASDVRGRLDEGQ--------------------------
MAV_4566|M.avium_104 STATNLGGREDVRTATAAPR
MMAR_0945|M.marinum_M NAAVHFGADADLMAHDQPG-
MLBr_02288|M.leprae_Br4923 --------------------
Mflv_0746|M.gilvum_PYR-GCK TAAAALGELELIYPWDTGPS
Mvan_0100|M.vanbaalenii_PYR-1 TAAAALGELELIYPRPATG-
MSMEG_5547|M.smegmatis_MC2_155 --------------------