For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LTHSRAVKAMDLLAVEHEVRQDTVVVRAKGDVDSSTVEQLVVHLAAAMQLAATHPARLVVIDLQPVTFFG SAGLNAVLECHEQGMAAGTSVRLVADHGQVLAPIQVTELDRIFEIYPTLAGALRPGKPGEHDLEDVERPR
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
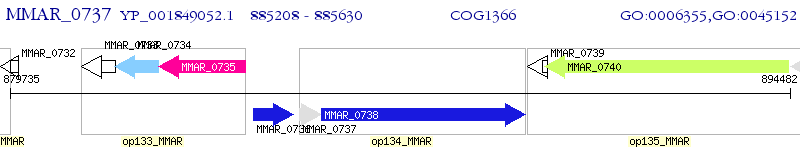
| M. marinum M | MMAR_0737 | - | - | 100% (140) | anti sigma factor antagonist |
| M. marinum M | MMAR_5181 | - | 3e-06 | 32.08% (106) | anti-anti-sigma regulatory factor |
| M. marinum M | MMAR_0214 | - | 8e-06 | 32.35% (102) | hypothetical protein MMAR_0214 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4424 | - | 1e-25 | 50.83% (120) | anti-sigma-factor antagonist |
| M. tuberculosis H37Rv | Rv3687c | rsfB | 3e-05 | 28.57% (126) | anti-anti-sigma factor RSFB (anti-sigma factor antagonist) |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. avium 104 | MAV_0435 | - | 7e-08 | 34.91% (106) | anti-anti-sigma factor |
| M. smegmatis MC2 155 | MSMEG_6127 | - | 2e-05 | 29.36% (109) | anti-anti-sigma factor |
| M. thermoresistible (build 8) | TH_1832 | - | 2e-12 | 39.66% (116) | ANTI-ANTI-SIGMA FACTOR RSFB (ANTI-SIGMA FACTOR ANTAGONIST) |
| M. ulcerans Agy99 | MUL_3602 | - | 3e-70 | 99.24% (131) | anti sigma factor antagonist |
| M. vanbaalenii PYR-1 | Mvan_1481 | - | 1e-13 | 37.96% (108) | anti-sigma-factor antagonist |
CLUSTAL 2.0.9 multiple sequence alignment
Rv3687c|M.tuberculosis_H37Rv ----MSAPDSITVTVADH--NGVAVLSIGGEIDLITAAALEEAIGEVVAD
MAV_0435|M.avium_104 -------------MVEDH--DGVSVVSVSGEIDLVTAPALEQAIGAVVAD
TH_1832|M.thermoresistible__bu --------------VEHR--DDSAVLRVQGVVDLATGDALQQAIDDLIAT
MSMEG_6127|M.smegmatis_MC2_155 ----MTSQDPANCTVEERRVGDITVVAVTGTVDMLTAPKLEDAIGSAAKS
Mvan_1481|M.vanbaalenii_PYR-1 -MDVSGHGRSRGFSVSPDRQGAALVLRVTGDLDAMTAPTLVTHLDIALAD
MMAR_0737|M.marinum_M MTHSRAVKAMDLLAVEHEVRQDTVVVRAKGDVDSSTVEQLVVHLAAAMQL
MUL_3602|M.ulcerans_Agy99 ---------MDLLAVEHEVRQDTVVVRAKGDVDSSTVEQLVVHLAAAMQL
Mflv_4424|M.gilvum_PYR-GCK -----MAERVGLLEIGQEVHDVAVVVRAGGEVDSGTVETLAAALDSAVAA
: *: * :* * * :
Rv3687c|M.tuberculosis_H37Rv NPT----ALVIDLSAVEFLGSVGLKILAATSEKIG-QSVKFGVVARGSVT
MAV_0435|M.avium_104 SPP----ALVIDLSAVEFLGSVGLKILAATYEKLG-KETGFGVVARGPAT
TH_1832|M.thermoresistible__bu RPS----ALVVDLSAVDFLASVGLQILVATHERLS-PSARFAVVADGPAT
MSMEG_6127|M.smegmatis_MC2_155 EPS----AVVVDLSAVDFLASAGMGVLVAAHGELA-PAVRLVVVADGPAT
Mvan_1481|M.vanbaalenii_PYR-1 APP----VVVVDMTDVEFLSSAGISVLVETHRLAERADISLRVVADGPAT
MMAR_0737|M.marinum_M AATHPARLVVIDLQPVTFFGSAGLNAVLECHEQGMAAGTSVRLVADHGQV
MUL_3602|M.ulcerans_Agy99 AATHPARLVVIDLQPVTFFGSAGLNAVLECHEQGMAAGTSVRLVADHGQL
Mflv_4424|M.gilvum_PYR-GCK AAAHPTRTVVVDLDGITYFGSAGLNALLTCREEGEGRGVVVRLVATTAEV
.. :*:*: : ::.*.*: : . :**
Rv3687c|M.tuberculosis_H37Rv RRPIHLMGLDKTFRLFSTLHDALTGVRGGRIDR-------
MAV_0435|M.avium_104 RRPIHLTGLDKTFPLYPTLDDALTAVRDGKLNG-------
TH_1832|M.thermoresistible__bu SRPIQLTRLDEVIALYPTLDEALSGVRTGERAE-------
MSMEG_6127|M.smegmatis_MC2_155 SRPLKLVGIADVVDLFATLDEALSSLKT------------
Mvan_1481|M.vanbaalenii_PYR-1 SRPFRMMRLDEVIDLYPTLADAMGERQQGRPPT-------
MMAR_0737|M.marinum_M LAPIQVTELDRIFEIYPTLAGALRPGKPGEHDLEDVERPR
MUL_3602|M.ulcerans_Agy99 LAPIQVTELDRIFEIYPTLAGALRPGKPGEHDLEDVERPR
Mflv_4424|M.gilvum_PYR-GCK TRPIEVTRLARILRPYPSVGEAVAAPE------DPPEDPR
*:.: : . :.:: *: .