For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MKFLQQMRKLFGLAAKFPARLTIAVIGTALLAGLVGVVGDTAIAVAFSKPGLPVEYLQVPSPSMGHDIKI QFQGGGQHAVYLLDGLRAQEDYNGWDINTPAFEEYYHSGLSVIMPVGGQSSFYSNWYQPSQGNGQHYTYK WETFLTQEMPSWLQANKNVLPTGNAAVGLSMSGSSALILASYYPQQFPYAASLSGFLNPSEGWWPTMIGL AMNDSGGYNANSMWGPSTDPAWKRNDPMVQIPRLVANNTRIWVYCGNGAPNELGGDNIPAKFLESLTLST NEIFQNTYAASGGRNGVFNFPPNGTHSWPYWNQQLVAMKPDIQQILNGSNNNA
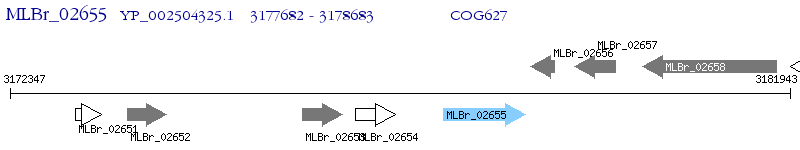
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. leprae Br4923 | MLBr_02655 | fbpC2 | - | 100% (333) | secreted antigen 85A, mycolyltransferase |
| M. leprae Br4923 | MLBr_00097 | fbpA | e-124 | 63.44% (331) | antigen 85A, mycolytransferase |
| M. leprae Br4923 | MLBr_02028 | fbpB | e-115 | 63.07% (306) | antigen 85A, mycolyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0134c | fbpC | 1e-167 | 82.07% (329) | secreted antigen 85-C FBPC (85C) (antigen 85 complex C) |
| M. gilvum PYR-GCK | Mflv_3335 | - | 1e-135 | 68.28% (331) | putative esterase |
| M. tuberculosis H37Rv | Rv0129c | fbpC | 1e-167 | 82.07% (329) | secreted antigen 85-C FBPC (85C) (antigen 85 complex C) |
| M. abscessus ATCC 19977 | MAB_0176 | - | 1e-107 | 54.85% (330) | antigen 85-A precursor |
| M. marinum M | MMAR_0328 | fbpC | 1e-168 | 82.93% (328) | secreted antigen 85-C FbpC |
| M. avium 104 | MAV_5183 | - | 1e-169 | 81.08% (333) | antigen 85-C |
| M. smegmatis MC2 155 | MSMEG_3580 | - | 1e-137 | 69.23% (325) | antigen 85-C |
| M. thermoresistible (build 8) | TH_2078 | fbpA | 1e-127 | 65.67% (335) | SECRETED ANTIGEN 85-A FBPA (MYCOLYL TRANSFERASE 85A) |
| M. ulcerans Agy99 | MUL_4793 | fbpC | 1e-168 | 82.93% (328) | secreted antigen 85-C FbpC |
| M. vanbaalenii PYR-1 | Mvan_3061 | - | 1e-134 | 67.67% (331) | putative esterase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3335|M.gilvum_PYR-GCK MKWKEKARG---TRKSLVRRVAAAALAAAALPGLIGVAGGSA---TAGAF
Mvan_3061|M.vanbaalenii_PYR-1 MKLKEKWRG---TARAAMRRVAAAALAAVALPGLIGFAGGSA---TAGAF
MSMEG_3580|M.smegmatis_MC2_155 MRGIAAWK--------ALRRFVIGALAALMLPGLIGFAGGSA---SAGAF
Mb0134c|M.bovis_AF2122/97 MTFFEQVRRLRSAATTLPRRLAIAAMGAVLVYGLVGTFGGPA---TAGAF
Rv0129c|M.tuberculosis_H37Rv MTFFEQVRRLRSAATTLPRRLAIAAMGAVLVYGLVGTFGGPA---TAGAF
MMAR_0328|M.marinum_M MKFVEKLR---GAAAGAPRRLTIAAIGAALLSGLVGAVGGAA---TAGAF
MUL_4793|M.ulcerans_Agy99 MKFVEKLR---GAAAGAPRRLTIAAIGAALLSGLVGAVGGAA---TAGAF
MLBr_02655|M.leprae_Br4923 MKFLQQMRKLFGLAAKFPARLTIAVIGTALLAGLVGVVGDTA---IAVAF
MAV_5183|M.avium_104 MSFIEKVRKLRGAAATMPRRLAIAAVGASLLSGVAVAAGGSP---VAGAF
TH_2078|M.thermoresistible__bu MKLVAKLRG---AARALPRRLTVAALAAALLPGLVGAFGGSA---TAGAF
MAB_0176|M.abscessus_ATCC_1997 MKLFSKMRG--ALARQSARRIAVAATAVAVLPGVAGIVGGTALTPVAGAF
* : *.. .. .. : *: *... * **
Mflv_3335|M.gilvum_PYR-GCK SRPGLPVEYLDVFSPSMNRDIRIQFQGGG--PHAVYLLDGLRAQEDYNGW
Mvan_3061|M.vanbaalenii_PYR-1 SRPGLPVEYLDVFSPSMGRDIRVQFQGGG--PRAVYLLDGLRAQEDYNGW
MSMEG_3580|M.smegmatis_MC2_155 SRPGLPVEYLDVFSPSMNRDIRVQFQGGG--PHAVYLLDGLRAQDDYNGW
Mb0134c|M.bovis_AF2122/97 SRPGLPVEYLQVPSASMGRDIKVQFQGGG--PHAVYLLDGLRAQDDYNGW
Rv0129c|M.tuberculosis_H37Rv SRPGLPVEYLQVPSASMGRDIKVQFQGGG--PHAVYLLDGLRAQDDYNGW
MMAR_0328|M.marinum_M SRPGLPVEYLQIPSAAMGRDIKVQFQGGG--AHSVYLLDGLRAQDDYNGW
MUL_4793|M.ulcerans_Agy99 SRPGLPVEYLQIPSAAMGRDIKVQFQGGG--AHSVYLLDGLRAQDDYNGW
MLBr_02655|M.leprae_Br4923 SKPGLPVEYLQVPSPSMGHDIKIQFQGGG--QHAVYLLDGLRAQEDYNGW
MAV_5183|M.avium_104 SKPGLPVEYLEVPSPSMGRNIKVQFQGGG--PHAVYLLDGLRAQDDYNGW
TH_2078|M.thermoresistible__bu SRPGLPVEYLMVPSAAMGRDIKVQFQSGGEGSPGVYLLDGLRARDDFNGW
MAB_0176|M.abscessus_ATCC_1997 SRPGLPVEYLQVPSASMGREIKVQFQPGG--AKAVYLLDGLRARDDFSGW
*:******** : *.:*.::*::*** ** .*********::*:.**
Mflv_3335|M.gilvum_PYR-GCK DINTPAFEWFYQSGLSTIMPVGGQSSFYSDWYQPSRGNGQNYTYKWETFL
Mvan_3061|M.vanbaalenii_PYR-1 DINTPAFEWFYQSGLSTIMPVGGQSSFYTDWYQPSRGNGQNYTYKWETFL
MSMEG_3580|M.smegmatis_MC2_155 DINTPAFEWFYQSGLSTIMPVGGQSSFYTDWYQPSKGNGQDYTYKWETFL
Mb0134c|M.bovis_AF2122/97 DINTPAFEEYYQSGLSVIMPVGGQSSFYTDWYQPSQSNGQNYTYKWETFL
Rv0129c|M.tuberculosis_H37Rv DINTPAFEEYYQSGLSVIMPVGGQSSFYTDWYQPSQSNGQNYTYKWETFL
MMAR_0328|M.marinum_M DINTPAFEEYYNSGLSVVMPVGGQSSFYSDWYQPSQGNGQGYTYKWETFL
MUL_4793|M.ulcerans_Agy99 DINTPAFEEYYNSGLSVVMPVGGQSSFYSDWYQPSQGNGQGYTYKWETFL
MLBr_02655|M.leprae_Br4923 DINTPAFEEYYHSGLSVIMPVGGQSSFYSNWYQPSQGNGQHYTYKWETFL
MAV_5183|M.avium_104 DINTPAFEEFYQSGLSVIMPVGGQSSFYSNWYQPSSGNGQNYTYKWETFL
TH_2078|M.thermoresistible__bu DIETAAFEWYYESGLSVIMPVGGQSSFYSDWYSPACGNNGCQTYKWETFL
MAB_0176|M.abscessus_ATCC_1997 DIETTAFEDYYQSGISMVMPVGGQSSFYTDWYNPAKGKDGVWTYKWETFT
**:*.*** :*.**:* :**********::**.*: .:. *******
Mflv_3335|M.gilvum_PYR-GCK TQELPAWLEANRGVSRTGNAVVGLSMAGSASLTYAIHHPQQFIYAASLSG
Mvan_3061|M.vanbaalenii_PYR-1 SQELPAWLEANRGVSRTGNAVVGLSMAGSAALTYAIYHPQQFIYAASLSG
MSMEG_3580|M.smegmatis_MC2_155 TQELPAWLEANRGVSRTGNAVVGLSMAGSAALTYAIYHPEQFIYASTLSG
Mb0134c|M.bovis_AF2122/97 TREMPAWLQANKGVSPTGNAAVGLSMSGGSALILAAYYPQQFPYAASLSG
Rv0129c|M.tuberculosis_H37Rv TREMPAWLQANKGVSPTGNAAVGLSMSGGSALILAAYYPQQFPYAASLSG
MMAR_0328|M.marinum_M TREMPAWLQANKGVSPTGNAAVGLSMSGGSALVLAAYYPQQFPYAASLSG
MUL_4793|M.ulcerans_Agy99 TREMPAWLQANKGVSPTGNAAVGLSMSGGSALVLAAYYPQQFPYAASLSG
MLBr_02655|M.leprae_Br4923 TQEMPSWLQANKNVLPTGNAAVGLSMSGSSALILASYYPQQFPYAASLSG
MAV_5183|M.avium_104 TQEMPLWMQSNKQVSPAGNAAVGLSMSGGSALILAAYYPQQFPYAASLSG
TH_2078|M.thermoresistible__bu TQELPAYLASNYGVDRNRNAAVGLSMAGSAALVLAIYYPQQFQYAASLSG
MAB_0176|M.abscessus_ATCC_1997 TQELPAYLAANKGISQTGNAVVGLSMGASAALTLSIYHPQLFVYAGALSG
::*:* :: :* : **.*****...::* : ::*: * **.:***
Mflv_3335|M.gilvum_PYR-GCK FLNPSEGWWPMLIGLAMNDAGGFNAESMWGPSSDP--AWKRNDPMVNINQ
Mvan_3061|M.vanbaalenii_PYR-1 FLNPSEGWWPMLIGLAMNDAGGFNAESMWGPSSDP--AWKRNDPMVNINQ
MSMEG_3580|M.smegmatis_MC2_155 FLNPSEGWWPMLIGLAMNDAGGYNAESMWGPSTDP--AWKRNDPMVNINQ
Mb0134c|M.bovis_AF2122/97 FLNPSEGWWPTLIGLAMNDSGGYNANSMWGPSSDP--AWKRNDPMVQIPR
Rv0129c|M.tuberculosis_H37Rv FLNPSEGWWPTLIGLAMNDSGGYNANSMWGPSSDP--AWKRNDPMVQIPR
MMAR_0328|M.marinum_M FLNPSEGWWPTLIGLAMNDSGGYNANSMWGPSTDP--AWKRNDPMVQIPR
MUL_4793|M.ulcerans_Agy99 FLNPSEGWWPTLIGLAMNDSGGYNANSMWGPSTDP--AWKRNDPMVQIPR
MLBr_02655|M.leprae_Br4923 FLNPSEGWWPTMIGLAMNDSGGYNANSMWGPSTDP--AWKRNDPMVQIPR
MAV_5183|M.avium_104 FLNPSEGWWPTLIGLAMNDSGGYNANSMWGPSTDP--AWKRNDPMVQIPR
TH_2078|M.thermoresistible__bu FLNLSEGWWPMLVGLAMGDAGGYDPNDMWGRTEDPNSAWKRNDPMVNIDK
MAB_0176|M.abscessus_ATCC_1997 FLNPSD--MKFQIGLAMGDAGGFSASDMWGPDSDP--AWQRNDPFLNIQK
*** *: :****.*:**:....*** ** **:****:::* :
Mflv_3335|M.gilvum_PYR-GCK LVANNTRIWIYCGTGTPSDLDSGTPGQNLMAAQFLEGFTLRTNVAFRDNY
Mvan_3061|M.vanbaalenii_PYR-1 LVANNTRIWIYCGTGTPSDLDNGTPGNNLMAAQFLEGFTLRTNIAFRDNY
MSMEG_3580|M.smegmatis_MC2_155 LVANNTRLWVYCGTGTPSELDAGVSGGNLLAAQFLEGLTLRTNLTFRDQY
Mb0134c|M.bovis_AF2122/97 LVANNTRIWVYCGNGTPSDL-----GGDNIPAKFLEGLTLRTNQTFRDTY
Rv0129c|M.tuberculosis_H37Rv LVANNTRIWVYCGNGTPSDL-----GGDNIPAKFLEGLTLRTNQTFRDTY
MMAR_0328|M.marinum_M LVANNTRIWVYCGNGTPSDL-----GGDNMPAKFLEGLTLRTNQTFRDTY
MUL_4793|M.ulcerans_Agy99 LVANNTRIWVYCGNGTPSDL-----GGDNMPAKFLEGLTLRTNQTFRDTY
MLBr_02655|M.leprae_Br4923 LVANNTRIWVYCGNGAPNEL-----GGDNIPAKFLESLTLSTNEIFQNTY
MAV_5183|M.avium_104 LVANNTRIWVYCGNGTPSDL-----GGDNVPAKFLEGLTLRTNEQFQNNY
TH_2078|M.thermoresistible__bu LVANNTRIWVFCGDGTPADIGAGVPEGDNFNAKFLESFTLRTNKTFQQEY
MAB_0176|M.abscessus_ATCC_1997 IIDNGTRLWIYCGTGDSTDLDATRNGFENFTGGFLEGMAIGSNKKFVEAY
:: *.**:*::** * . :: : . . ***.::: :* * : *
Mflv_3335|M.gilvum_PYR-GCK IAAGGNNGVFNFPTSGTHSWGYWGQQLQQMKPDIQRVLGAQATA------
Mvan_3061|M.vanbaalenii_PYR-1 IAAGGTNGVFNFPTSGTHSWGYWGQQLQQMKPDIQRVLGAQATV------
MSMEG_3580|M.smegmatis_MC2_155 IAAGGTNAVFNFPPNGTHTWNYWGQQLLEMKPDIQRVLGAQSAT------
Mb0134c|M.bovis_AF2122/97 AADGGRNGVFNFPPNGTHSWPYWNEQLVAMKADIQHVLNGATPPAAPAAP
Rv0129c|M.tuberculosis_H37Rv AADGGRNGVFNFPPNGTHSWPYWNEQLVAMKADIQHVLNGATPPAAPAAP
MMAR_0328|M.marinum_M LASGGRNGVFNFPTNGTHSWPYWNQQLVAMKGDILKVLNGPAVPAAPAAP
MUL_4793|M.ulcerans_Agy99 LASGGRNGVFNFPTNGTHSWPYWNQQLVAMKGDILKVLNGPAVPAAPAAP
MLBr_02655|M.leprae_Br4923 AASGGRNGVFNFPPNGTHSWPYWNQQLVAMKPDIQQILNGSNNNA-----
MAV_5183|M.avium_104 AAAGGRNGVFNFPANGTHSWPYWNQQLMAMKPDMQQVLLSGNTTAAPAQP
TH_2078|M.thermoresistible__bu IAAGGKNGVFNFPDAGTHSWGYWGQQLQQMKPDIQRVLGVSAA-------
MAB_0176|M.abscessus_ATCC_1997 TAAGGKNAHIEFPPGGIHNWTYWGQQLRAMKPDMVAYLQSH---------
* ** *. ::** * *.* **.:** ** *: *
Mflv_3335|M.gilvum_PYR-GCK -----------------
Mvan_3061|M.vanbaalenii_PYR-1 -----------------
MSMEG_3580|M.smegmatis_MC2_155 -----------------
Mb0134c|M.bovis_AF2122/97 AA---------------
Rv0129c|M.tuberculosis_H37Rv AA---------------
MMAR_0328|M.marinum_M EAPAAPVAPAA------
MUL_4793|M.ulcerans_Agy99 EAPAAPVAPAA------
MLBr_02655|M.leprae_Br4923 -----------------
MAV_5183|M.avium_104 AQPAQPAQPAQPAQPAT
TH_2078|M.thermoresistible__bu -----------------
MAB_0176|M.abscessus_ATCC_1997 -----------------